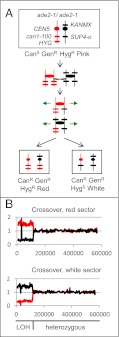
Fig. 2.
Detection of reciprocal crossovers leading to LOH events. (A) Pattern of marker segregation resulting from a reciprocal crossover. At the top of the panel, we show the arrangements of markers on chromosome V in the diploid strain SLA11.7. The chromosome is shown as a vertical line, the location of the markers by short horizontal lines, and the centromere by circles; red and black signify that the chromosomes were derived from the W303a-related haploid and the YJM789-related haploid, respectively. As discussed in the text, the SUP4-o–encoded tRNA partially suppresses the red colony phenotype associated with the ade2-1 mutation and suppresses the canavanine-resistance phenotype associated with can1-100. A reciprocal crossover event can produce two canavanine-resistant daughter cells. One daughter cell is canavanine-resistant and forms a red sector because it lacks the SUP4-o gene, and the other is canavanine-resistant because it lacks any CAN1 gene and forms a white sector because it has two copies of SUP4-o. Both sectors retain one copy of the KANMX gene and thus are geneticin-resistant. (B) Analysis of genomic DNA isolated from red and white sectors resulting from a reciprocal crossover (sectored colony PG311-16B; ref. 13). Genomic DNA was purified from each sector of a canavanine-resistant red/white -sectored colony and examined by microarrays capable of distinguishing LOH at a resolution of 1 kb throughout the genome. We measured the normalized hybridization of genomic DNA from the experimental strains and the control diploid to oligonucleotides with SNPs specific to W303a or YJM789 (the haploid strains used to construct the diploid). The y-axis shows the ratio of hybridization (experimental/control) for SNP-specific oligonucleotides, with red and black lines indicating hybridization to W303a- and YJM789-specific oligonucleotides, respectively. The x-axis shows Saccharomyces Genome Database (SGD) coordinates. The red sector has an LOH event in which W303a-derived SNPs become homozygous, and the white sector has the reciprocal LOH event. This pattern is consistent with a reciprocal crossover occurring near SGD coordinate 120,000. The “spike” of hybridization near SGD coordinate 30,000 (Fig. 3B) is an artifact reflecting the substitution of the can1 gene with SUP4-o (13).