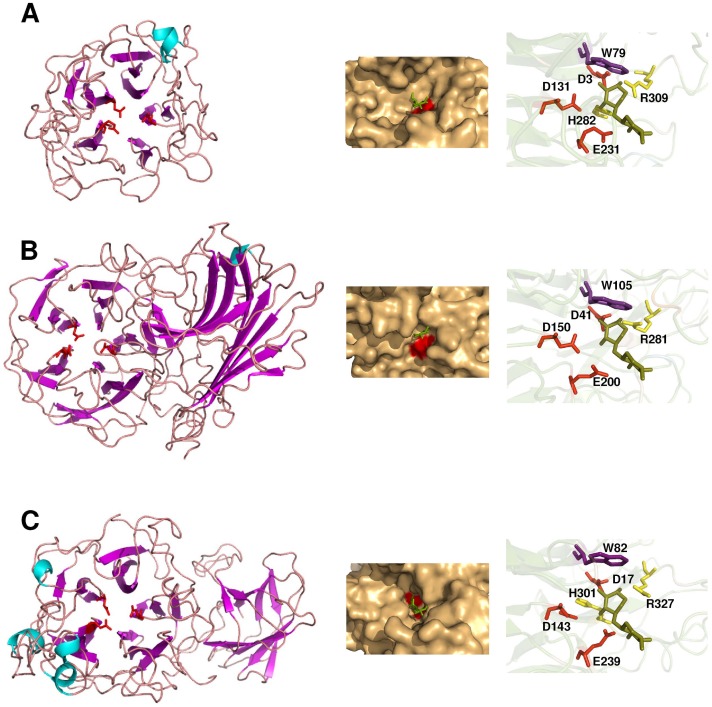
Figure 3. Structural models of R-03_04 (A), R-03_05 (B) and R-09_02 (C).
The putative catalytic residues (general acid, base and transition state stabilisers) are depicted in red. The left panel shows the overall folding of the protein. The right panel is a close-up view of the catalytic site, indicating the catalytic residues and other highly conserved residues among the GHF43 enzymes that may establish polar (yellow) or hydrophobic (violet) contacts with the substrate at the −1 subsite. The middle panel shows the solvent-accessible surface close to the catalytic site. As a reference, the location of a xylobiose molecule (green) is given according to the structural superimposition with the β-xylosidase from Geobacillus stearothermophilus (PDB code 2EXJ).