Figure 4.
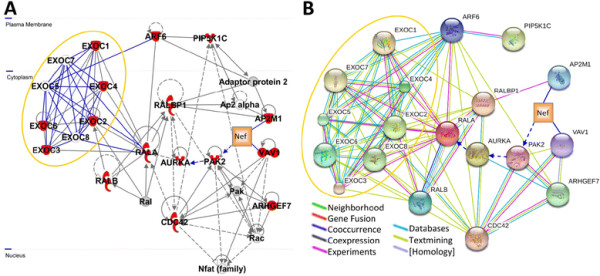
Visualization of protein interaction networks of Nef-interacting proteins and exocyst complex components, regulators, and effectors.A) IPA network of Nef-interacting proteins and exocyst complex components, regulators, and effectors. Inputting 16 molecules (depicted as red nodes, described in the text) into Ingenuity Pathways Analysis [Ingenuity® Systems, www.ingenuity.com] yielded a network of 35 molecules, of which 13 [Calmodulin, Dock1-Pak, ERK1/2, Exocyst, Jnk, NFkB (complex), Phosphatidylinositol 4,5 kinase, PI3K (complex), PIP4K2C, Pkc(s) Pld, Ras, and TCR] were omitted for clarity. Components of the exocyst complex are encircled by a gold oval. Manually-curated edges are shown in dark blue, and dashed arrows denote activation via indirect association. IPA assigned this network a score of 42, which is based on the hypergeometric distribution and is the -log(right-tailed Fisher’s exact test result). The score implies a 1 in 1042 chance of obtaining a network containing at least the same number of network eligible molecules by chance when randomly picking 16 molecules that are present in networks from the Ingenuity Knowledge Base. B) STRING network of Nef-interacting proteins and exocyst complex components, regulators, and effectors. Inputting 19 molecules (16 molecules input into IPA, plus EXOC5, EXOC7, and EXOC8; depicted as spheres) into STRING yielded a network visualizing linkages between Nef, the exocyst complex, and regulators of the actin cytoskeleton. Components of the exocyst complex are encircled by a gold oval. Manually-curated edges are shown in dark blue, and dashed arrows denote activation via indirect association. Other edge colors reflect the evidence channels (see legend) by which protein nodes are related in the STRING database. Scores STRING assigned to relationships depicted in this network are provided in Additional file 4: Table S3.
