Abstract
We characterized three phases of Hyacinthus orientalis L. embryo sac development, in which the transcriptional activity of the cells differed using immunolocalization of incorporated 5′-bromouracil, the total RNA polymerase II pool and the hypo- (initiation) and hyperphosphorylated (elongation) forms of RNA Pol II. The first stage, which lasts from the multinuclear stage to cellularization, is a period of high transcriptional activity, probably related to the maturation of female gametophyte cells. The second stage, encompassing the period of embryo sac maturity and the progamic phase, involves the transcriptional silencing of cells that will soon undergo fusion with male gametes. During this period in the hyacinth egg cell, there are almost no newly formed transcripts, and only a small pool of RNA Pol II is present in the nucleus. The transcriptional activity of the central cell is only slightly higher than that observed in the egg cell. The post-fertilization stage is related to the transcriptional activation of the zygote and the primary endosperm cell. The rapid increase in the pool of newly formed transcripts in these cells is accompanied by an increase in the pool of RNA Pol II, and the pattern of enzyme distribution in the zygote nucleus is similar to that observed in the somatic cells of the ovule. Our data, together with the earlier results of Pięciński et al. (2008), indicate post-fertilization synthesis and the maturation of numerous mRNA transcripts, suggesting that fertilization in H. orientalis induces the activation of the zygote and endosperm genomes.
Keywords: Embryo sac, Egg cell, Zygote, Fertilization, Transcription, RNA polymerase II (RNA Pol II)
Introduction
The structure and development of the female gametophyte and the processes of fertilization and early embryo development have been described quite well in many species of angiosperms (Huang and Russell 1992; Russel 1996; Higashiyama 2002, 2003). The embryo sac in approximately 70% of angiosperm species, including Arabidopsis thaliana and Zea mays, develops according to the Polygonum type (among others, Huang and Russell 1992; Drews et al. 1998). During megasporogenesis four haploid megaspores are formed from a diploid megaspore mother cell, of which three undergo degeneration. The remaining so-called functional megaspore undergoes three mitotic divisions, which are not accompanied by cytokinesis. These divisions lead to the formation of an embryo sac with eight nuclei in which cytokinesis and cell differentiation take place. Finally, three of the four nuclei of the chalazal region become the nuclei of the antipodal cells, two nuclei of the micropylar region become synergid nuclei, and the third nucleus of the region becomes the nucleus of the egg cell. Two nuclei are the so-called polar nuclei, which undergo fusion, forming a diplohaploid nucleus of the central cell. The egg cell and the central cell participate in the process of double fertilization, which is unique to flowering plants. Following fusion with one of the sperm cells, the egg cell forms a zygote, which produces a new sporophyte generation, while the fertilized central cell develops into the nutritive endosperm tissue.
The nuclei of the embryo sac are probably characterized by a distinct pattern of gene expression, and the developing cells acquire unique properties related to their different biological functions because of this pattern. Results in recent years, mainly in Z. mays and A. thaliana, have provided information about the level of expression of some selected genes in the cells of the embryo sac (Dresselhaus et al. 1999a, b; Cordts et al. 2001; Márton et al. 2005; Lê et al. 2005; Johnston et al. 2007; Wuest et al. 2010). Several thousand genes are estimated to be potentially involved in the development of the embryo sac in Arabidopsis. Among the 1,260 analysed genes, only 8.6% of the transcripts were not found in sporophyte tissues, indicating their specific expression in the cells of the female gametophyte (Johnston et al. 2007). In the Arabidopsis (Steffen et al. 2007; Wuest et al. 2010), maize (Lê et al. 2005; Yang et al. 2006) and wheat (Sprunck et al. 2005) egg cell, the expression of specific genes has been noted but still our knowledge of the time course of the genetic differentiation of the sister nuclei in the embryo sac is still incomplete.
In contrast to oocytes and animal embryos, several problems have still not been explained in flowering plants, including (1) transcripts storage in the egg cell, (2) the regulation of the early stages of embryonic development, and (3) the time of activation of the zygotic genome. Investigations are usually performed in vitro (Scholten et al. 2002) or only in certain species of plants, mainly in Z. mays and A. thaliana, often due to technical problems with whole unfertilized and fertilized ovules (Grimanelli et al. 2005). The results of the genetic analysis of the egg cell, the zygote and the early stages of embryonic development are intriguing. It is postulated as in animals (Telford et al. 1990) that the early stages of embryonic development in plants are controlled by maternal transcripts, and the paternal genome remains silenced after fertilization (Vielle-Calzada et al. 2000; Grimanelli et al. 2005; Pillot et al. 2010). In fact, it has been shown that the A. thaliana zygote remains relatively quiescent and that the embryo can undergo several divisions in the absence of the de novo transcription, thus relying on deposited maternal products (Pillot et al. 2010). However, other studies in this species have shown that several alleles from both the maternal and paternal genomes are transcribed shortly after fertilization (Weijers et al. 2001; Köhler et al. 2005; Autran et al. 2011). Early in vitro experiments in Z. mays indicated that the expression of numerous genes increases in the zygote, including those involved in replication and those encoding ribosomal proteins (Dresselhaus et al. 1999a, b, 2005). In the fertilized egg cell, the genes of cell cycle regulation are also transcribed de novo (Sauter et al. 1998). This result indicated an earlier activation of the zygote genome and post-fertilization gene expression in processes such as the replication and cell cycle regulation of the fertilized Z. mays egg cell. Similar results were obtained for Nicotiana (Ning et al. 2006). The expression of 9 gene homologues was identified in the Nicotiana zygote in vitro, but transcripts of these genes were not present in the egg cell. This result proved that in tobacco the activation of the zygote genome has already taken place prior to its division (Ning et al. 2006). Also, Zhao et al. (2011) using expression profiling analysis of egg cells, zygotes and two-celled proembryos in tobacco indicated that maternal-to-zygotic transition is initiated prior to the first zygotic division and de novo transcripts appearing in later zygotes are probably required to trigger embryogenesis. Further investigations in Zea, using transgenic sperm cells containing green fluorescent protein (GFP), demonstrated the capability of maize zygotes for the de novo gene expression 4 h after fertilization when a decondensation of sperm chromatin has already taken place (Scholten et al. 2002). In turn, the analysis of the allele-specific expression of 25 genes after fertilization of the egg cell in maize showed immediate equivalent paternal genomic contribution to the zygote. Also, each gene expressed before the first cell division of the zygotes showed paternal transcripts, demonstrating distinct transcriptional activation of the paternal genome (Meyer and Scholten 2007). Thus, the presented studies indicate that in Z. mays the zygote is transcriptionally active, and the activation of the zygotic genome occurs directly after fertilization. These results are in contrast to the results obtained by Grimanelli et al. (2005). Genetic analysis of fertilized and unfertilized maize embryos indicates that only maternally inherited alleles are expressed for 3 days following fertilization. These data suggest that the transition from the maternal to the zygotic regulation of embryo development takes place many days after fertilization.
Taken together, all studies show that different regulation strategies may be present for embryonic development in higher plants. In some species, the activation of zygotic genes occurs directly after fertilization and these transcripts play a key role in the development of the embryo and endosperm (Ray 1997; Pagnussat et al. 2005). In others, as in animals, the expression of the zygotic genome is not required for the first divisions of the embryo and takes place at different stages of its development.
The transcription of protein-coding genes is a multi-stage process performed by RNA polymerase II. This enzyme is composed of 12 protein subunits (Lee and Young 2000). The largest subunit has a unique region at its C-terminus designated the CTD (Carboxy-Terminal Domain). A characteristic property of the CTD is the presence of several dozen tandem repeats of a sequence composed of seven amino acids: Tyr-Ser-Pro-Thr-Ser-Pro-Ser. This motif is highly conserved in Eukaryota (Corden et al. 1985) and determines the CTD activity, which is regulated by the degree of phosphorylation of the serine residues at positions 2 and 5 (Hahn 2004). The form with low levels of phosphorylation (Pol IIA) is linked with the preinitiation complex, in which the serine residue at position 5 is phosphorylated. In a transcriptionally active complex, the CTD is characterized by the presence of an additional phosphate group at position 2. The hyperphosphorylated form of RNA Pol II (Pol IIO) is thought to be responsible for elongation (Komarnitsky et al. 2000). Moreover, the Ser2P of the CTD is responsible for the interaction of the transcriptional apparatus with the machinery responsible for polyadenylation and pre-mRNA maturation (Proudfoot et al. 2002). At the end of a round of transcription, a dephosphorylation of the CTD generally takes place, which makes the breakdown and recycling of the transcriptional machinery possible (Kamenski et al. 2004).
Determining the levels of different types of RNA Pol II may be an important indication of gene expression in the cells of fertilized and unfertilized embryo sacs. So far, this technique has only been used in A. thaliana (Pillot et al. 2010). Differences in the levels of the active forms of RNA Pol II, which were positively correlated with their different transcriptional activities, were observed between the nuclei of the embryo sac. In the transcriptionally silenced zygote, the level of the hyperphosphorylated form of RNA Pol II was very low, in contrast to endosperm nuclei, in which gene expression was high.
Our investigations in Hyacinthus orientalis using immunocytochemical methods and in situ hybridization are aimed at determining the transcriptional activity and the type of RNA synthesized in vivo in the cells of developing embryo sacs, both unfertilized and fertilized. We have previously shown that, while the egg cell of H. orientalis does not accumulate large amounts of poly(A) RNA, a slightly larger pool of this RNA is present in the central cell (Pięciński et al. 2008). A very high level of poly(A) RNA is, however, present in the zygote. Thus, in this species, de novo synthesis of polyadenylated transcripts takes place shortly after fertilization. The accumulation of poly(A) RNA was accompanied by a rapid increase in the level of splicing factors, indicating that the synthesis and maturation of numerous mRNA transcripts take place in the nucleus of the hyacinth zygote (Pięciński et al. 2008).
The aim of the present investigations was to determine in vivo the total transcriptional activity of cells of the H. orientalis embryo sac during successive stages of its functioning, i.e., from the nuclear period up to fertilization. We also analysed the progamic phase, which had not been investigated thus far. This phase encompasses the period of growth of the pollen tube in the transmission pathway of the pistil when the embryo sac is preparing to accept the male gametophyte for fertilization. The evaluation of the total transcriptional activity of the hyacinth female gametophyte cells was performed by analysing the level and distribution of newly formed transcripts using the immunolocalization of incorporated 5′-bromouracil. To better understand which types of RNA are transcribed in embryo sac cells, the levels of the total pool of RNA Pol II and of the hypo- and hyperphosphorylated forms of RNA Pol II were also analysed, along with the levels and distribution of pre-rRNA and rRNA. The experiments allowed the evaluation of the levels of newly formed transcripts and RNA Pol II in the cells of the embryo sac and a comparison with the levels of these molecules in the somatic cells of the ovule.
Materials and methods
Plant material
Hyacinthus orientalis L. variety Pink Pearl ovules were used in the investigation. Plants were obtained from a commercial cultivar and grown at room temperature in the Institute of General and Molecular Biology, Nicolaus Copernicus University, Toruń, Poland. Ovules were mechanically isolated from flower buds (±5–10 mm), mature, unpollinated flowers (after the opening of the flower) and 4 h, 8 h (progamic phase—when pollen tubes have reached about three-quarters of the style length and did not enter the female gametophyte) and 96 h (fertilized embryo sacs) after hand cross-pollinated flowers. The growth of the pollen tubes was checked: isolated pistils were cut up and placed in 0.01% aniline blue and were examined in fluorescence microscopy.
Localization of transcription
BrU incorporation was performed according to Smoliński et al. (2007). Directly after isolation, ovules were incubated in 0.01% water solution of Triton X-100 (Fluka) for 10 min. Then the material was washed three times for 10 min with distilled water and incubated in 20 mM water solution of 5′-bromouracil (BrU, Sigma) for 1 h in room temperature. The long incubation allowed BrU to penetrate through all the layers of the ovule and enter the embryo sac cells. It facilitated the comparison of transcripts at all analysed stages, which differed in RNA synthesis intensity. Under these conditions, part of the product moved from the synthesis site to the cytoplasm at some stages. Ovules were then washed three times for 10 min with PBS buffer, pH 7.2. The chemical fixation, embedding in butyl methyl methacrylate (BMM, Fluka) and semithin sectioning (Leica Ultracut) were performed as previously described (Pięciński et al. 2008). Incorporated 5′-bromouracil was detected by incubating with a primary, mouse-anti-BrdUTP antibody (Sigma) in 1% bovine serum albumin (BSA) (Sigma-Aldrich) in PBS buffer, pH 7.2 (1:200), overnight at 4°C, and the secondary, goat-anti-mouse antibody Alexa Fluor 488 (Invitrogen) in 1% BSA in PBS buffer, pH 7.2 (1:1,000), for 1 h at 37°C. Control reactions were performed without the primary antibody. DNA was stained with 4,6-diamidino-2-phenylindole (DAPI; Fluka).
Semithin sections were observed in Nikon Eclipse 80i fluorescence microscope. The CPI Plan Fluor 100× (N.A. 1.3) DIC H/N2 immersion oil lens and narrow band filters (UV-2EC, B-2EC, G-2EC) were used. The results were registered with a Nikon DS-5Mc colour cooled digital camera and Lucia General 5.00 software (Laboratory Imaging, Prague, Czech Republic).
Immunolabelling of RNA polymerase II
For light microscopy, immunolocalization of total RNA Pol II pool and hypo- and hyperphosphorylated forms of the RNA Pol II ovules were prepared as previously described (Pięciński et al. 2008). After blocking with 0.01% ac BSA (Sigma) in PBS buffer, pH 7.2 semithin section was incubated with primary IgG mouse monoclonal anti-RNA Pol II CTD 4H8 (recognizes the total RNA Pol II pool) or two kinds of primary IgM mouse monoclonal antibodies: anti-RNA polymerase II H14 (recognizes the phosphoserine 5 version of RNA Pol II, Pol IIA) and RNA polymerase II H5 (recognizes the phosphoserine 2 version of RNA Pol II, Pol IIO) (Covance) in 0.01% ac BSA in PBS buffer, pH 7.2 (1:500), overnight at 4°C. Antibodies binding was detected with the goat, anti-mouse antibody Alexa Fluor 488 (Invitrogen) in 0.01% ac BSA in PBS buffer, pH 7.2 (1:1000). Incubation was carried out for 1 h at 37°C. In the control incubations with the primary antibodies were omitted. DNA was stained with DAPI (Fluka). Semithin sections were analysed with a Olympus BX50 fluorescence microscope. The UPlanFI 100× (N.A. 1.3) oil immersion lens and narrow band filters (U-MNU, U-MNG) were used. The results were registered with a Olympus XC50 digital colour camera and CellB software (Olympus Soft Imaging Solutions GmbH, Münster, Germany).
For immunogold labelling of RNA Pol II ovules were fixed in 4% paraformaldehyde and 0.25% glutaraldehyde in PBS buffer, pH 7.2, for 24 h at 4°C. Material were washed, dehydrated in graded concentrations of ethanol up to 100% and embedded in LR Gold (Sigma). Samples polymerization LR Gold with 1% benzoyl peroxide as the accelerator occurred for 3 days at −20°C. Ultrathin sections were cut using a Leica UTC ultramicrotome, collected on the nickel grids coated with 0.3% formvar (Sigma). After blocking for 15 min with 3% BSA in PBS buffer, pH 7.2 sections were treated with anti-RNA polymerase II 4H8, H14, H5 primary antibodies (Covance) in 0.2% ac BSA in PBS buffer, pH 7.2 (1:50). Antibody binding was detected by incubation with 15 nm diameter gold conjugated goat, anti-mouse antibody (BB International) diluted in 0.2% ac BSA in PBS buffer, pH 7.2 (1:30) for 1 h at 37°C. Controls were performed by omitting incubation with the primary antibody. The sections were stained with 1% phosphotungstic acid and 5% uranyl acetate solutions and examined using a JEOL 1010 transmission electron microscope at 80 kV.
Quantitative evaluations
Image analysis was performed on serial semithin sections after immunofluorescence staining (BrU, total RNA Pol II pool), with each reaction step performed using consistent values of temperature, incubation times, concentration of primary and secondary antibodies. Quantitative analysis of fluorescence intensity for immunofluorescence staining was carried out for 5–7 each cell types (5 sections per cell) from each development stage. All measurements were conducted at the same magnification, field area (controlled with a shutter), and positioning of the fibre optics cable. Camera settings were kept constant for exposition time, gain and offset. Lucia G software was used to determine the average μm3 signal intensity of each studied cell compartment and is expressed in a.u. (arbitrary unit of fluorescence intensity). For all antigens and developmental stages, the obtained data were corrected for background autofluorescence as determined from negative control signal intensities.
Ten images for each antigen (total RNA Pol II pool, RNA Pol IIO, RNA Pol IIA) were taken for gold grains analysis. The area of nucleus was measured using the NIS-Elements AR3.00 software (Nikon). The number of gold particles was counted, and the labelling density for each nucleus was expressed as the number of gold particles per μm2.
To test differences among multiple samples (groups, i.e., antigen level in different stages), a Kruskal–Wallis ANOVA test was used. Statistical data and graphs were created using Microsoft Excel 2007 software.
Results
Localization of transcription
The pattern of the distribution of newly formed transcripts in the H. orientalis embryo sac in the period from the nuclear stage to the fertilization was determined. The investigations were not quantitative but allowed for the evaluation of the levels of newly formed transcripts and RNA Pol II in the cells of the embryo sac. The levels of these molecules were compared with the levels observed in the somatic tissues of the ovule.
Ovule
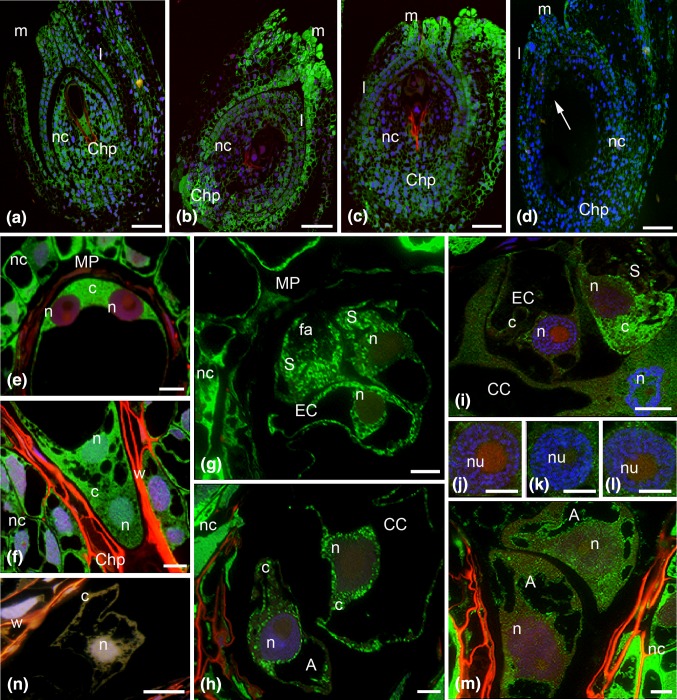
In immature ovules, a strong signal, localized in the 4-nucleate embryo sac, was similar to that which occurred in somatic cells (Fig. 1a). There were no significant differences in signal between the different tissues of the ovule, i.e., the fluorescence levels of the nucellus and the integument cells were similar. After the cellularization process of the embryo sac, before the opening of the flower, a differentiation of the signal strength between gametophyte cells and somatic cells was observed in the ovule (Fig. 1b). In ovules analysed using a small microscope magnification, the fluorescence of the embryo sac was almost invisible. In this period, differences in the labelling of transcripts were observed in the somatic tissues of the ovule. The labelling of the nucellus cells was lower than before cellularization, whereas the signal in the integument cells increased. A high fluorescence was localized in the cells of the micropyle and the chalazal region of the ovule. A particularly high level of newly formed transcripts was observed in the cells of the integument surrounding the micropylar canal of the ovule (Fig. 2l). In these cells, the high fluorescence was observed both in the cytoplasm and in the nucleus. In the area of the micropyle, strong labelling of the cytoplasm was observed in cells of the nucellus. The level of the fluorescence in nuclei was significantly lower. A similar pattern of labelling of newly formed transcripts was present in the mature ovule, isolated from an opened flower (Fig. 1c). In somatic tissues, an intense signal was localized in cells surrounding the micropyle and in cells of the funiculus found near the micropyle. The labelling of the embryo sac was lower than in the surrounding nucellus cells. This signal pattern was also maintained during the progamic phase (not shown). In the ovule, into which the pollen tube had grown and the fertilization had taken place, a distinct decrease in the labelling of newly formed transcripts in cells of the surrounding micropyle was observed (Fig. 1d). In the large embryo sac, a weak fluorescence of the zygote and endosperm was visible.
Fig. 1.
Immunofluorescence localization of incorporated 5′-bromouracil of the H. orientalis ovule a with the 4-nucleate embryo sac, b after cellularization of the embryo sac, c mature ovule at anthesis, d after fertilization and embryo sac (e, f) 4-nucleate; young 7-celled, before anthesis: g egg apparatus cells, h central and antipodal cells; mature embryo sac: i egg cell, synergid and central cells, (j–l) the next sections of the egg cell nucleus, m antipodal cells; n control reaction. A antipodal cell, c cytoplasm, CC central cell, ChP chalazal pole, EC egg cell, fa filiform apparatus, I integument cells, m micropyle, MP micropylar pole, n nucleus; nc nucellus cells, nu nucleolus, S synergid, w embryo sac wall, green newly formed transcripts, blue DAPI staining. Scale bars are 100 μm (a–d), 10 μm (e, f, j, l), 20 μm (g–i, m, n)
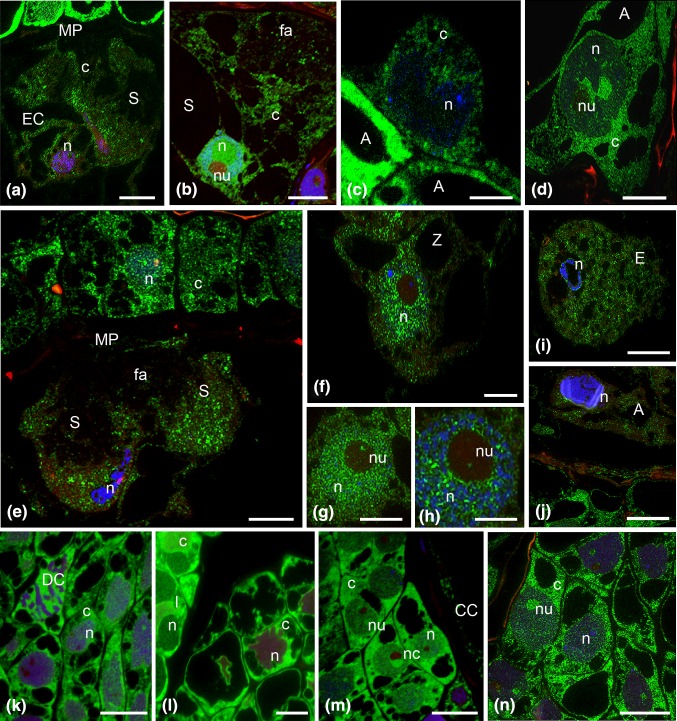
Fig. 2.
Immunofluorescence localization of incorporated 5′-bromouracil of the H. orientalis embryo sac, during the progamic phase: egg cell and synergid cell (a, b), central cell (c), the antipodal cell (d); fertilized embryo sac: degenerated synergid cells with condensed chromatin (e), zygote nucleus (f), the next sections of the zygote nucleus (g, h), endosperm (i), the degenerated antipodal cell (j); somatic cells of the ovule: with 4-nucleate embryo sac (k), with young 7-celled embryo sac (l), during the progamic phase (m) and after fertilization (n). A antipodal cell, c cytoplasm, CC central cell, DC divided cell, E endosperm cell, EC egg cell, fa filiform apparatus, I integument cells, MP micropylar pole, n nucleus, nc nucellus cells, nu nucleolus, S synergid, Z zygote, green newly formed transcripts, blue DAPI staining. Scale bars are 10 μm (a–j) and 20 μm (k–n)
Embryo sac
The labelling of newly formed transcripts in the 4-nucleate embryo sac was high and comparable to the labelling of the nucellus cells (Fig. 1e, f). In the micropylar region of the embryo sac, fluorescence that indicated the presence of newly formed transcripts was predominantly localized in the cytoplasm (Fig. 1e). The signal observed there was similar to that which occurred in the surrounding somatic cells. In contrast, the signal in the nuclei was very low. In the chalazal region of the embryo sac, the signal was localized both in the cytoplasm as well as in cell nuclei (Fig. 1f). The strength of the nuclear signal was higher than or similar to that occurring in the nuclei of somatic cells. During this period of ovule development, a very high signal was observed in nucellus cells (Fig. 2k). The labelling of newly formed transcripts was localized both in the nuclei, including the nucleoli, as well as in the cytoplasm.
After cellularization relatively strong fluorescence that indicated newly formed transcripts was still observed in the cells of the embryo sac (Fig. 1g, h). The signal present there was only slightly weaker than in the surrounding nucellus cells. In the cells of the newly formed egg apparatus, the distribution and strength of the signal were comparable between synergids and the egg cell (Fig. 1g). In the egg cell, newly formed transcripts were localized mainly in the cytoplasm, with the exception of the vacuoles. The strength of nuclear labelling was much lower. Synergid cells were characterized by a similar intensity and distribution of green fluorescence (Fig. 1g). A relatively high, uniform signal was present throughout the entire non-vacuolar cytoplasm, and the strength of the fluorescence was weaker only around the filiform apparatus. In the nuclei of the synergid cells, the fluorescence was distinctly lower. In the strongly vacuolarised central cell the signal was similar to that which was observed in the cells of the egg apparatus (Fig. 1h). Newly formed transcripts were mainly present in the cytoplasm. The nuclear signal was weaker, and the labelling was observed in the form of numerous small fluorescent spots. In the antipodal cells the fluorescence was localized in the cytoplasm, and the nuclear signal was very low (Fig. 1h).
In the mature embryo sac of H. orientalis, significant differences in the labelling of its individual cells were observed. With the exception of the synergid cells, the level of newly formed transcripts in the embryo sac cells was significantly lower than that observed in the surrounding somatic cells (Fig. 1i, m). In the egg apparatus, newly formed transcripts were observed in the synergid cells (Fig. 1i). Within them, the highest signal occurred in the perinuclear cytoplasm and in the basal region. Towards the apical pole, the level of the signal decreased; however, foci of fluorescence also occurred around the filiform apparatus. The labelling of the synergid nuclei was clearly lower. The fluorescence signal in the egg cell was significantly weaker than that in the synergid cells (Fig. 1i). A low signal was present in the cytoplasm, and the labelling of the nucleus was almost invisible. Only single, diffusely dispersed spots of fluorescence were observed in the nucleoplasm and the nucleolus (Fig. 1j–l). In the central cell, the labelling of the newly formed transcripts was higher than in the egg cell (Fig. 1i). The fluorescence of the nucleus was very weak. A relatively weak, diffusely dispersed signal was localized mainly in the cytoplasm. Slightly higher labelling was observed only in the cytoplasm around the nucleus. The cells of the embryo sac in which the highest labelling was observed were the antipodal cells (Fig. 1m). The fluorescence of these cells was present in the nucleus, including the nucleoli, and in the cytoplasm.
After pollination of the pistil in cells targeted by sperm cells, a low level of newly formed transcripts was still observed. In the egg cell, the signal was localized in the cytoplasm with only a few spots of fluorescence visible in the nucleus (Fig. 2a). In the central cell, the labelling was higher than in the egg cell, similar to the pattern observed before pollination (Fig. 2c). Numerous fluorescence spots were observed in the cytoplasm. Foci of very weak, dispersed fluorescence were also observed in the nucleus. Significant changes in labelling of newly formed transcripts were observed in synergid cells. In comparison to the period before the pollination of the pistil, the fluorescence of these cells was lower and was only slightly higher than in the egg cell in most analysed embryo sacs (Fig. 2a). Single fluorescence spots were localized predominantly in the perinuclear cytoplasm. However, in some embryo sacs, the labelling in the synergid cells was still relatively high. Single fluorescence spots were present both in the cytoplasm as well as in the nucleus and nucleolus (Fig. 2b). In this period of embryo sac development, a relatively high level of newly formed transcripts was observed in the antipodal cells (Fig. 2d). Fluorescence was observed both in the cytoplasm and in the nucleus of these cells. A strong fluorescence, much higher than that observed in the cells of the embryo sac, was found in the cells surrounding the embryo sac (Fig. 2m). An intense signal was localized in the cytoplasm and in the nuclei of nucellus cells. The nuclear signal was generally slightly weaker than that observed in the cytoplasm. In the nuclei, a relatively high accumulation of signal was observed in the nucleoli.
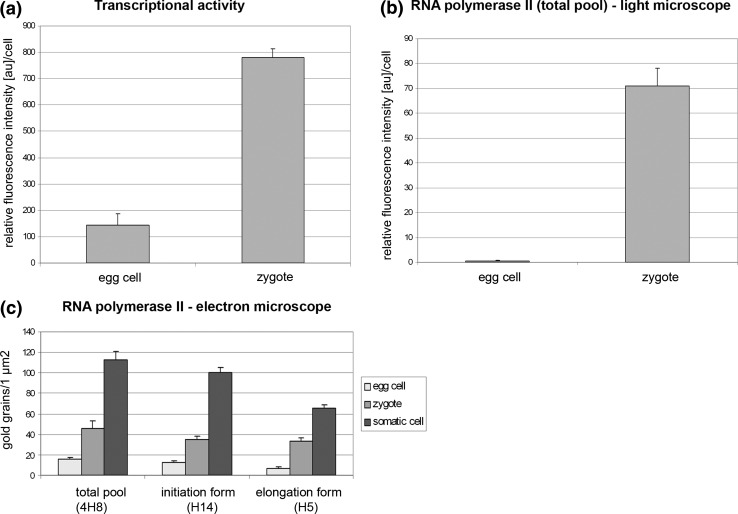
After fertilization, changes in the distribution and in the level of newly formed transcripts were observed in the cells of the embryo sac. The labelling patterns of synergid cells within the same egg apparatus were different (Fig. 2e). In the cell degenerating after the growth of the pollen tube, fluorescence was distinctly lower than in the synergid cell not penetrated by the male gametophyte. In the fertilized embryo sac, very high labelling of newly formed transcripts occurred in the zygote (Fig. 2f). The level of fluorescence was much higher than in egg cell before fertilization and increased about fivefold (Fig. 6a). A signal in the form of smaller and greater of fluorescence spots was present both in the nucleus and the cytoplasm. The labelling in the nucleus was higher than that in the cytoplasm. In this early period of zygote development, the fluorescence was almost completely absent in the nucleolus (Fig. 2g, h). On successive sections through the nucleolus, a strong signal was observed in the form of single fluorescence spots, probably around the chromatin of the nuclear organizer. Only individual small foci of signal were visible outside this area. A high level of newly formed transcripts was also observed in the primary endosperm cell (Fig. 2i). Numerous small fluorescence foci were generally localized in the cytoplasm, and single spots of signal were also visible in the nucleus. In antipodal cells, which showed signs of degeneration (strong condensation of chromatin, occasional fragmentation of nuclei), the level of newly synthesized transcripts was very low (Fig. 2j). A weak signal was present mainly in the cytoplasm. In nuclei with strongly condensed chromatin, only single fluorescence spots were observed. After fertilization, a relatively high labelling of newly synthesized transcripts was observed in the cells surrounding an embryo sac that was still increasing in size (Fig. 2n). Fluorescence was localized both within the nuclei and the cytoplasm, but the cytoplasmic signal was slightly higher than the nuclear one, in general. In these cells, very strong labelling was also observed in the nucleoli.
Fig. 6.
Histogram illustrating the intensities of incorporated 5′-bromouracil (a) and the total RNA Pol II pool fluorescence (b) in egg cell before and after fertilization. c Graph comparing the levels (gold grains) of the total RNA Pol II pool, hypophosphorylated (initiation) and hyperphosphorylated (elongation) forms of RNA Pol II between the nucleus of egg cell, zygote and somatic cell
No labelling of the nuclei and cytoplasm of the embryo sac cells was observed in a control reaction (Fig. 1n).
Distribution of RNA polymerase II
Immunofluorescence analysis made it possible to determine the level of RNA polymerase II in the female gametophyte before and after fertilization in relation to the level of this enzyme in somatic cells of the ovule (Fig. 3).
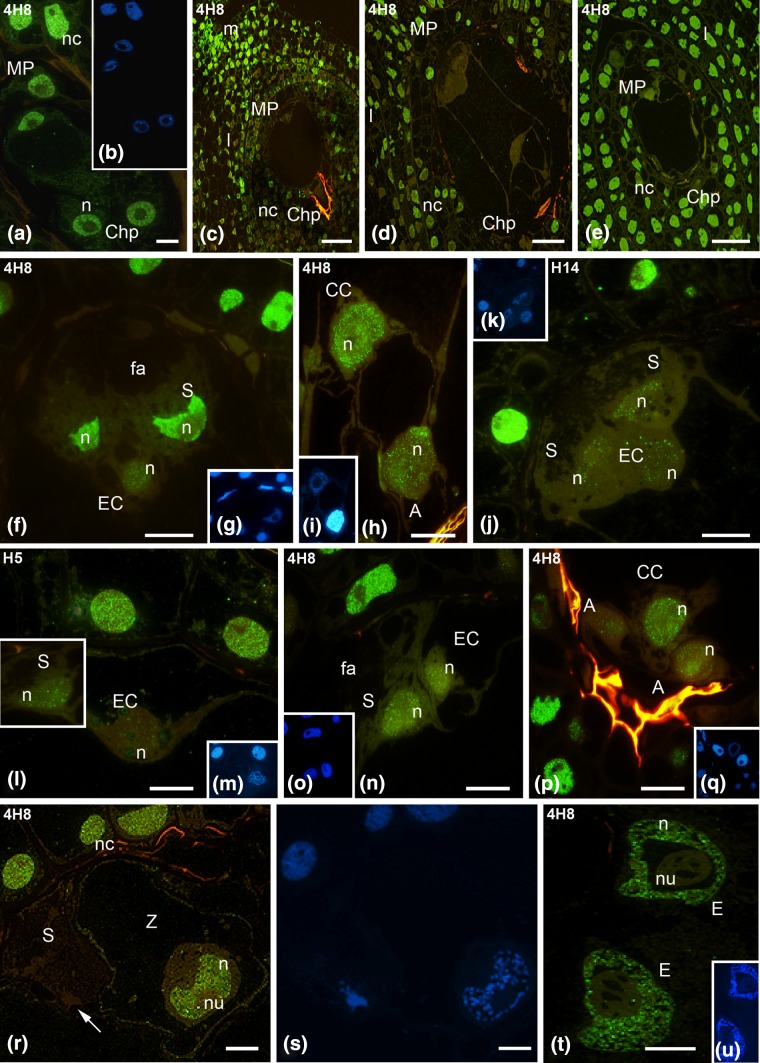
Fig. 3.
Immunofluorescence localization of the total Pol RNA II pool (4H8 antibodies) and the hypophosphorylated (Pol IIA, H14 antibodies) and the hyperphosphorylated (Pol RNA IIO, H5 antibodies) forms of RNA Pol II of the H. orientalis ovule and embryo sac. 4-Nucleate embryo sac (a), mature ovule with the embryo sac after cellularization (c), ovule during the progamic phase (d), ovule after fertilization (e). Mature embryo sac: egg apparatus (f), (h) central and antipodal cells (h), egg apparatus (j–l); embryo sac during the progamic phase: micropylar region (n), chalazal region (p). Embryo sac after fertilization: zygote and degenerated synergid cell (r), endosperm nuclei (t). DAPI staining (b, g, i, k, m, o, q, s, u). m Micropyle, I integument cells, nc nucellus cells, MP micropylar pole, ChP chalazal pole, S synergid, EC egg cell, CC central cell, A antipodal cell, fa filiform apparatus, n nucleus, nu nucleolus, c cytoplasm, green RNA Pol II, blue DAPI staining. Scale bars 10 μm (a, f, h, j–l, n, p, r), 100 μm (c), 50 μm (d, e)
At the nucleate stage of the gametophyte, the signal indicating the presence of RNA Pol II was only slightly lower than that in the nuclei of somatic cells (Fig. 3a). In all nuclei of the embryo sac, both the distribution and the strength of the signal were similar. In the mature ovule, a clear differentiation of the fluorescence of the embryo sac and somatic cells was observed (Fig. 3c). Using a small microscope magnification, the labelling of the cells of the embryo sac was almost invisible. In somatic tissues, the highest signal was observed in the nuclei of the integument cells surrounding the micropyle, while a lower fluorescence was observed in the nuclei of the nucellus cells. A similar pattern of RNA Pol II labelling was present during the progamic phase. The signal from the nuclei of the embryo sac was very weak; however, the signal was observed in the nuclei of somatic cells (Fig. 3d). At this level of analysis, a weak fluorescence of the zygote and endosperm was only visible in the embryo sac after fertilization (Fig. 3e).
The cells of the mature embryo sac differed in the level of the total pool of RNA Pol II (Fig. 3f, h). In the egg apparatus, a high level of labelling, only slightly lower than in the nucellus cells, was observed in the synergid cells (Fig. 3f). RNA Pol II was evenly distributed in the extranucleolar nucleoplasm. The labelling of the egg cell nucleus was weaker. The signal was present in the area of the whole nucleus in the form of numerous clusters (Fig. 3f). The labelling of RNA Pol II in the nucleus of the central cell was clearly higher than in the nucleus of the egg cell (Fig. 3h). The signal occurred in the form of numerous clusters of fluorescence, which were present in the whole nucleoplasm. The fluorescence of the antipodal cell nuclei was slightly lower than that of the central cell nucleus. The labelling of these nuclei was observed in the form of numerous clusters of fluorescence irregularly distributed in the area of the nucleoplasm (Fig. 3h). The labelling of the egg apparatus cells with H14 and H5 antibodies showed the presence of both forms of RNA Pol II in the nuclei of the synergid cells and the egg cell. The RNA Pol IIA signal (the hypophosphorylated form of RNA Pol II) was observed in the form of fluorescence foci, whose number was similar in all nuclei of the egg apparatus (Fig. 3j). In contrast, the treatment of these cells with H5 antibody, which labels the hyperphosphorylated RNA Pol II form (Pol IIO), revealed a different localization of RNA Pol IIO in the nucleus of the egg cell and in synergid cells nuclei (Fig. 3l). The signal in the nucleus of the egg cell was very low, and only individual, small fluorescence spots were observed in this nucleus. The fluorescence of the synergid cell nucleus was distinctly higher, even though it was lower than the level of fluorescence observed in somatic cells.
During the progamic phase, the labelling of the total RNA Pol II pool was similar to that observed before pollination of the pistil in the nuclei of the egg cell and the central cell (Fig. 3n, p). In the nuclei of the egg cell (Fig. 3n) and central cell (Fig. 3p), small fluorescence spots were observed. A significantly decrease in labelling was observed in the nuclei of synergid cells (Fig. 3n). The strength of the signal was similar to that which was found in the egg cell. A decrease of the fluorescence intensity of the total RNA Pol II pool was also observed in the antipodal cells (Fig. 3p).
Significant changes in the distribution of the total RNA Pol II pool in the embryo sac cells of H. orientalis were observed after fertilization (Fig. 3r, t). The highest level of labelling was observed in the zygote nucleus (Fig. 3r). The signal from the zygote nucleus was comparable to the signal present in the nuclei of somatic cells of the ovule. The fluorescence in the zygote nucleus was evenly distributed throughout the entire nucleoplasm, outside the area of the nucleolus. The level of total RNA Pol II pool was significantly higher than in the egg cell before fertilization. The intensities of the fluorescence over a hundred fold increased (Fig. 6b). A high level of RNA Pol II labelling was also observed in the nuclei of the forming endosperm (Fig. 3t). After fertilization, the level of RNA Pol II in synergid cells was very low (Fig. 3r). In the nuclei of degenerating cells, the fluorescence signal was observed in some nuclei in the form of single foci, while other nuclei were completely unlabelled. Similarly, only a few spots of RNA Pol II fluorescence were observed in the antipodal cells (not shown).
Ultrastructural localization of RNA polymerase II
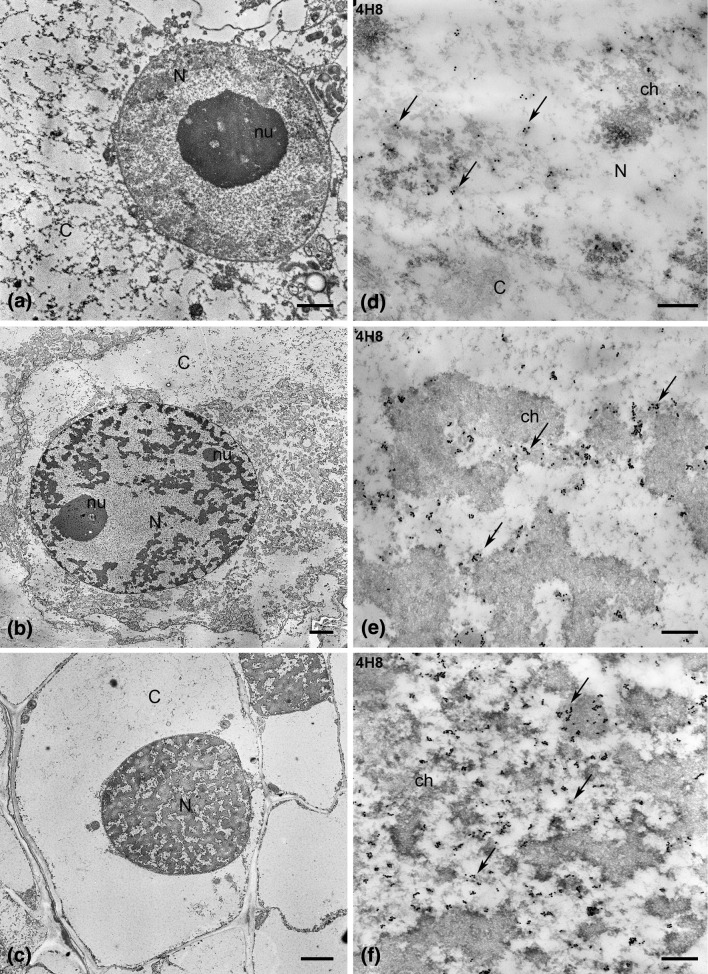
The levels of the RNA Pol II pools were observed in unfertilized and fertilized egg cells using electron microscopy. The pattern of labelling in the egg cell nucleus before and after fertilization was compared with the signal pattern that occur in the nuclei of somatic cells. Ultrastructural analysis revealed a different chromatin organization in the nuclei of the analysed cells. The nuclei of H. orientalis somatic cells had a typical reticulate structure (Fig. 4c). Chromatin in the egg cell nucleus, however, was highly decondensed (Fig. 4a). In the zygote, changes in chromatin organization took place. In a nucleus in which a second nucleolus is already visible, chromatin condensed and occurred in the form of larger and smaller aggregates (Fig. 4b).
Fig. 4.
Ultrastructure of the egg cell (a), zygote (b) and somatic cell (c) nuclei. Bar 2 μm. Immunogold localization of the total RNA Pol II pool (4H8 antibodies) in the egg cell (d), zygote (e) and somatic cell (f) of H. orientalis. Bar 200 nm. C cytoplasm, ch chromatin, N nucleus, nu nucleolus
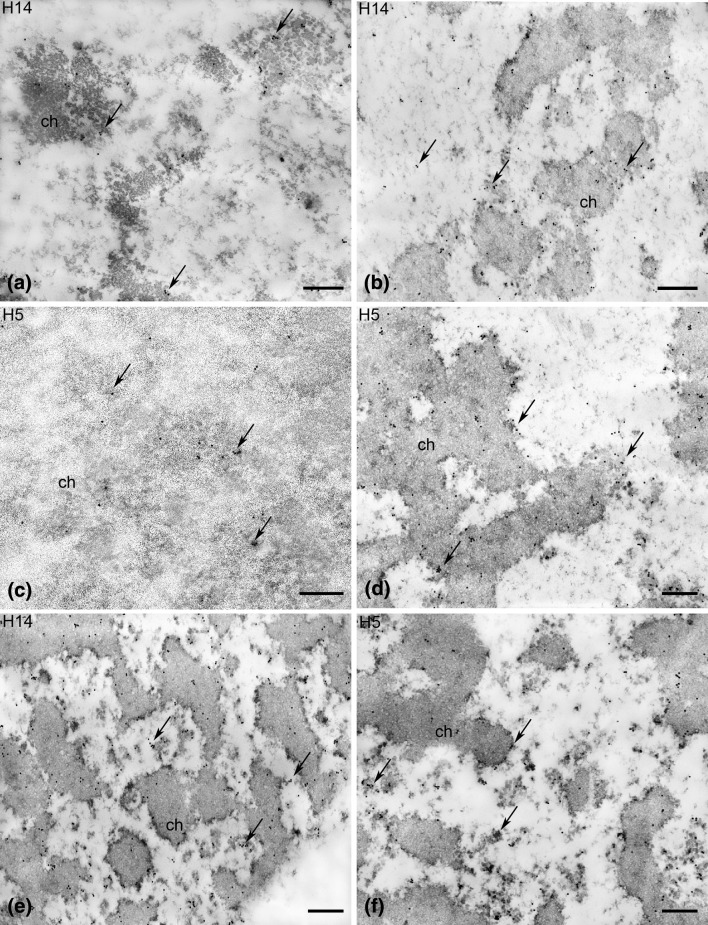
In the egg cell, RNA Pol II was observed in loose chromatin and in interchromatin areas (Fig. 4d). The number of gold grains in the egg nucleus was distinctly lower than in the somatic cell nucleus (Figs. 4f, 6c). The labelling of the zygote nucleus was distinctly higher than that of the egg nucleus before fertilization (Figs. 4e, 6c). The signal was localized mainly in perichromatin areas, and the gold grains were less numerous in interchromatin areas. A similar pattern of labelling was observed in the somatic cell nucleus (Fig. 4f). In the egg nucleus, a few gold grains showing the binding of H14 antibodies were observed inside and between loose chromatin and in the margins (Fig. 5a). The labelling with the H5 antibody was very weak (Fig. 6c); only individual gold grains were observed in loose chromatin (Fig. 5c). In the zygote nucleus, the signal showing the binding of H14 and H5 antibodies increased strongly (Fig. 6c). Gold grains indicating the presence of Pol IIA (Fig. 5b) and Pol IIO (Fig. 5d) were mainly observed in the perichromatin areas and within chromatin aggregates. Fewer gold grains were observed between chromatin aggregates, especially after labelling with the H14 antibody (Fig. 5b). The pattern of labelling with the H14 and H5 antibodies observed in the zygote nucleus was similar to that which was found in the somatic cell (Fig. 5e, f).
Fig. 5.
Immunogold localization of the hypophosphorylated (Pol IIA, H14 antibodies) and the hyperphosphorylated (Pol RNA IIO, H5 antibodies) forms of RNA Pol II in egg cell (a, c), zygote (b, d) and somatic cell (e, f) nuclei of the H. orientalis. ch chromatin. bar 200 nm
Discussion
Our investigations indicated that during the development of the H. orientalis ovule, changes in the transcriptional activity of somatic tissues and female gametophyte cells occurred. In the period between the cellularization of the embryo sac and the progamic phase, particularly high transcriptional activity is detected in the cells surrounding the micropyle, including the cells of the integument and funiculus. Their activity is silenced only after fertilization. The micropyle is the site through which the pollen tube grows into the embryo sac. The increased activity of cells around the micropylar canal is probably related to the synthesis of factors participating in the attraction of the pollen tube and its direction to the embryo sac (Huang and Russell 1992; Higashiyama et al. 2003; Yadegari and Drews 2004; Márton et al. 2005; Dresselhaus and Márton 2009).
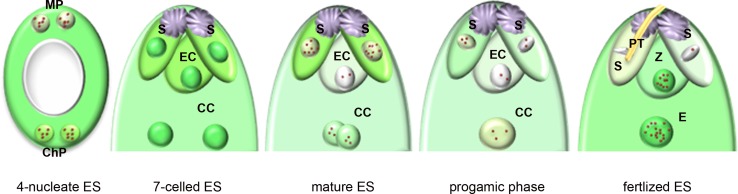
Upon in vivo analysis of the embryo sac, three developmental stages that differed in transcriptional activity were distinguished: (i) a stage of high transcriptional activity that lasted from the multinucleate stage until cellularization, (ii) a stage of transcriptional silencing encompassing the period of flower maturity and the progamic phase and (iii) the post-fertilization stage, which is related to the transcriptional activation of the zygote and endosperm (Fig. 7). In the free nucleate stage, the transcriptional activity of the embryo sac was high, similar to that which was observed in the somatic cells of the ovule. However, a different distribution of newly formed transcripts could already be observed between the poles of the embryo sac at the 4-nucleate stage. In the micropylar region, the newly formed transcripts were predominantly localized in the cytoplasm, whereas these transcripts were observed both in the nucleus and the cytoplasm in the chalazal region. Within each nucleus of the embryo sac, a similarly high level of RNA Pol II pool was present (Fig. 7). This results suggests that mRNA synthesis occurs at a similar level in these nuclei, and the observed differential transcript localization may be due to different rates of mRNA transport to the cytoplasm. It is possible that the different pattern of localization of newly synthesized RNA in the embryo sac regions reflects the differentiation in the metabolism of the nuclei, which already execute different differentiation programmes at this early stage. In the micropylar pole region, differentiation will ultimately lead to the formation of the egg apparatus, whereas antipodal cells will form in the chalazal region. A relatively high pool of newly formed transcripts was also observed in all seven cells directly after cellularization of the embryo sac (Fig. 7). The levels of newly formed transcripts in all cells of the female gametophyte were similar, but already lower than in the somatic cells of the ovule. The small differences in the level of transcriptional activity indicate that the maturation of female gametophyte cells lasts until the opening of the flower.
Fig. 7.
Transcriptional activity of Hyacinthus orientalis L. female gametophyte cells before and after fertilization. CC central cell, ChP chalazal pole, EC egg cell, ES embryo sac, MP micropylar pole, PT pollen tube, S synergid cell, Z zygote, green level of newly formed transcripts, red spots RNA Pol II
In the mature hyacinth embryo sac, i.e., after the opening of the flower, the transcriptional activity of its cells was distinctly lower than that of somatic cells. Distinct differences were also observed between the cells of the embryo sac. In the H. orientalis egg cell, the level of newly formed transcripts was low (Fig. 7). RNA Pol II was present in its nucleus; however, the pool of this enzyme in the female gamete was lower than in the otherscells of the embryo sac. Predominantly, the hypophosphorylated form of RNA Pol II (Pol IIA) was present. The pool of the hyperphosphorylated form of RNA Pol II (Pol IIO) was very low, indicating that mRNA synthesis is very limited in the mature egg cell. This results is in agreement with our earlier investigations, which indicated that the nucleus of the mature H. orientalis egg cell is almost completely devoid of the snRNAs that are indispensable for the maturation of primary mRNA transcripts (Pięciński et al. 2008). Thus, in H. orientalis, the egg cell is transcriptionally active during the period from the cellularization of the embryo sac to the opening of the flower. During anthesis, when the pistil becomes able to accept the male gametophyte, a transcriptional silencing of the female gamete takes place, including the inhibition of the transcription and maturation of mRNA. The amount of poly(A) RNA in the mature H. orientalis egg cell is also very low (Pięciński et al. 2008), which in turn indicates that this cell does not accumulate large amounts of transcripts synthesized during its maturation. Thus, the observed phenomenon is different from that observed in the animal egg cell, which stores maternal poly(A) RNA transcripts that participate in the control of the early stages of development (Picton et al. 1998).
The low transcriptional activity of the egg cell nucleus is consistent with the degree of condensation of its chromatin. Ultrastructural investigations have shown that the chromatin is highly decondensed in the hyacinth egg cell nucleus, similar to other analysed species (Diboll 1968; Coimbra and Salema 1999). It can be suggested that the loosening of chromatin unaccompanied by high transcriptional activity is the preparation of the nuclear genetic material for fusion with the chromatin of the sperm cell. To date, this hypothesis has not been the subject of any investigations and there is no evidence to confirm it.
Analysis of the transcriptional activity of the female gamete of flowering plants has only been performed in several species of plants, including Zea, Triticum, Nicotiana, and Arabidopsis, mainly by in vitro techniques. The obtained results indicated that the main transcripts present in the egg cell are related to basal metabolism, such as transcripts encoding proteins participating in replication, regulation of translation (Dresselhaus et al. 1999a, 1999b) and ribosome formation (Dresselhaus et al. 1999a; Sprunck et al. 2005; Ning et al. 2006), along with histone proteins (Sprunck et al. 2005) and signal proteins (Sprunck et al. 2005; Ning et al. 2006). After fertilization, the levels of most of these transcripts were significantly lower, suggesting that their presence is mainly related to the functioning of the egg cell, including its interaction with the male gamete and fertilization.
Our in vivo investigations also allowed us to determine the transcriptional activity of the female gamete also in the progamic phase, which had not been previously analysed. We found that when pollen tubes grew into the style of the pistil, the transcriptional activity of the hyacinth egg cell remained low, even though RNA Pol II was present in the nucleus (Fig. 7). At this stage of development, the female gamete has a low level of splicing factors and poly(A) RNA (Pięciński et al. 2008), which indicates its low activity in mRNA synthesis. Thus, in H. orientalis, the progamic phase is a period of low transcriptional activity of the egg cell that will soon undergo fusion with the sperm cell.
Significant changes in the levels of newly formed transcripts and RNA Pol II in the egg cell occured only after fertilization (Fig. 7). In the zygote, the pool of transcripts, which were mainly located in the extranucleolar areas, increased rapidly. The RNA Pol II pool that was present in the egg cell nucleus during the progamic phase probably participated in this process. We cannot exclude the possibility that, in H. orientalis, RNA Pol II activation occurs through changes in the degree of its phosphorylation, similar to the process described during early embryogenesis in animals (Bellier et al. 1997a, b; Palancade et al. 2001). However, in H. orientalis, high transcriptional activity of the zygote was also accompanied by an increase in the RNA Pol II pool (Fig. 7). This result indicates de novo synthesis of this enzyme after fertilization. In the zygote nucleus, the pattern of the level and distribution of RNA Pol II became similar to that observed in the somatic cells of the ovule. Both the initiation and the elongation forms of RNA Pol II were observed mainly at the margins of chromatin aggregates. In animals, this area is considered to be the main site of mRNA transcription and maturation (Cmarko et al. 1999; Niedojadło et al. 2011). In the H. orientalis zygote nucleus, the levels of snRNA and poly(A) RNA also increase (Pięciński et al. 2008). Our investigations of H. orientalis sperm cells have shown that mature male gametes lack splicing factors and RNA Pol II (Zienkiewicz et al. 2011) and, therefore do not contribute these molecules to target cells. Presumably, the transcription and maturation not only the new mRNA pool, but also of snRNA take place in the zygote. The above-described investigations, therefore, indicate that the activation of the zygote genome, including genes encoding proteins, snRNA and paternal rRNA, has already occurred before the first division of the zygote in hyacinth, in which the egg cell does not store large amounts of poly(A) RNA (Pięciński et al. 2008).
The time of the maternal-to-zygotic transition and the activation of the paternal genome in flowering plants remain open questions, as the results obtained thus far are unsolved. On the one hand, it was shown that, in contrast to H. orientalis, the A. thaliana zygote has a low level of transcriptional activity and a small pool of the elongation form of RNA Pol II in its nucleus (Pillot et al. 2010). Transcripts detected during the early stages of development of an embryo of this species are maternal transcripts (Vielle-Calzada et al. 2000; Grimanelli et al. 2005; Autran et al. 2011), and the zygote may divide several times before resuming transcription (Pillot et al. 2010). On the other hand, the investigations of Weijers et al. (2001) and Köhler et al. (2005) have shown that the transcription of several alleles from both the maternal and paternal genomes takes place in the A. thaliana zygote, which indicates the activation of the zygotic genome directly after fertilization. An early post-fertilization activation of the zygotic genome was also found in other analysed species, including Nicotiana (Ning et al. 2006, Zhao et al. 2011) and Zea (Scholten et al. 2002; Meyer and Scholten 2007). In turn, the genetic analysis of maize ovules performed by Grimanelli et al. (2005) indicated that only the maternal alleles are expressed during the early stages of embryo development.
The post-fertilization resumption of transcriptional activity is accompanied by changes in the organization of chromatin, which assumes a more condensed form than the egg cell chromatin. In the nucleus with this ultrastructure, two nucleoli are already visible, indicating the activation of the paternal NOR (nucleolar organizer region). In maize, karyogamy begins approximately 12 h after pollination, and two nucleoli are visible for 5–8 h (Mól et al. 1994), even though the paternal chromatin undergoes decondensation during the first 3 h after the fusion of the egg cell and the sperm cell (Scholten et al. 2002). Based on these data, we believe that the aggregates of condensed chromatin observed in the nucleus of the H. orientalis zygote are already formed after the dispersion of the sperm chromatin and are a transitional stage towards the organization of a reticulate type nucleus, which is characteristic for H. orientalis.
The transcriptional activity of the mature central cell was higher than that of the egg cell. A higher pool of newly formed transcripts and a relatively high level of RNA Pol II were observed in the central cell both after opening of the flower and during the progamic phase. The central cell also contains more splicing factors and poly(A) RNA than the egg cell (Pięciński et al. 2008). After fertilization, strong increase in the pool of newly synthesized transcripts occurred in the primary endosperm cell, indicating an early activation of the endosperm genome. The high transcriptional activity of endosperm nuclei was accompanied by a high level of RNA Pol II and an increase in the amount of splicing factors and poly(A) RNA (Pięciński et al. 2008). This result indicate that mRNA constitutes a large part of the newly synthesized transcripts. In the endosperm cell, these transcripts were localized mainly in the area of the cytoplasm, their levels of new transcripts in the nucleus were much lower. We cannot exclude the possibility that the distribution of new transcripts, which is different than that in the zygote, is due to the earlier initiation of endosperm development. Transcripts synthesized directly after the activation of the endosperm genome are rapidly transported to the cytoplasm, where they participate in the synthesis of proteins necessary for the development of the nutritive tissue. It is known that endosperm development has already begun prior to zygote division in many species of flowering plants, including H. orientalis (Yadegari and Drews 2004). The early activation of the parental genome during endosperm development does not arouse controversy. This process has also been observed in other plant species, including A. thaliana (Grimanelli et al. 2005; Pillot et al. 2010) and Z. mays (Scholten et al. 2002).
In contrast to the target cells for male gametes, the synergid cells of the mature egg apparatus of H. orientalis have high transcriptional activity. A rich pool of RNA Pol II and splicing factors is present in their nuclei (Pięciński et al. 2008). The high level of newly formed transcripts was localized predominantly in the cytoplasm, in which poly(A) RNA is also present (Pięciński et al. 2008). These results indicate that in the synergid cells of the mature embryo sac, mRNA among others molecules undergoes intense synthesis and maturation and is then transported to the cytoplasm. The high transcriptional activity observed in the synergid cells during anthesis is in agreement with the function of these cells. In this period of receptivity of the flower, the synergid cells synthesize and secrete attractants, which attract pollen tubes (Higashiyama et al. 2001; Weterings and Russell 2004; Márton et al. 2005). During the progamic phase, progressive silencing of the synergid transcriptional activity takes place. This silencing is shown by the decrease in the levels of newly formed transcripts, the RNA Pol II pool, splicing factors and poly(A) RNA (Pięciński et al. 2008). The synergid cells degenerated after fertilization, the fulfilment of their biological functions. This degeneration was accompanied by further inhibition of the transcriptional activity and the elimination of RNA pol II from the synergid cell nuclei.
In the mature hyacinth embryo sac and during the progamic phase, a higher level of transcriptional activity was observed in the antipodal cells, which play a nutritive role. Similar to the synergid cells, the metabolism of the degenerating antipodal cells decreased after fertilization, which was accompanied by a decrease in newly formed transcripts and the total RNA Pol II pool.
In conclusion, our earlier (Pięciński et al. 2008) and present investigations have revealed several characteristics of embryonic development in H. orientalis. First, the mature egg cell is almost completely transcriptionally silenced. Only a small pool of RNA Pol II and splicing factors is present in its nucleus, and it does not accumulate large amounts of poly(A) RNA. Second, the transcriptional activity of the central cell is slightly higher. Third, the transcriptional silencing of the cells that will soon undergo fertilization remains through the progamic phase. Finally, fertilization induces the activation of the zygote and primary endosperm cell genomes (Fig. 7). The mechanisms involved in the regulation of transcription and then translation immediately following the fertilization of both cells are not yet known.
Acknowledgments
We thank M. Świdziński for technical assistance. This project was supported by the Ministry of Science and Higher Education in Poland, grant No N N303 290434.
Glossary
- BrU
Bromouridine
- BSA
Bovine serum albumin
- PBS
Phosphate-buffered saline
- RNA Pol IIA
Hypophosphorylated (initiation) form of RNA Pol II
- RNA Pol IIO
Hyperphosphorylated (elongation) form of RNA Pol II
References
- Autran D, Baroux C, Raissig MT, Lenormand T, Wittig M, Grob S, Steimer A, Barann M, Klostermeier UC, Leblanc O, Vielle-Calzada JP, Rosenstiel P, Grimanelli D, Grossniklaus U. Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell. 2011;145:707–719. doi: 10.1016/j.cell.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Bellier S, Dubois MF, Nishida E, Almouzni G, Bensaude O. Phosphorylation of the RNA polymerase II largest subunit during Xenopus laevis oocyte maturation. Mol Cell Biol. 1997;17:1434–1440. doi: 10.1128/mcb.17.3.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellier S, Chastant S, Adenot P, Vincent M, Renard JP, Bensaude O. Nuclear translocation and carboxyl-terminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos. EMBO J. 1997;16:6250–6262. doi: 10.1093/emboj/16.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmarko D, Verschure PJ, Martin TE, Dahmus ME, Krause S, Fu XD, Driel R, Fakan S. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol Biol Cell. 1999;10:211–223. doi: 10.1091/mbc.10.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coimbra S, Salema R. Ultrastructure of the embryo sac development in Amaranthus hypochondriacus L. Ann Bot. 1999;84:781–789. doi: 10.1006/anbo.1999.0978. [DOI] [Google Scholar]
- Corden JL, Cadena DL, Ahearn JM, Jr, Dahmus ME. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci USA. 1985;82:7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordts S, Bantin J, Wittich PE, Kranz E, Lörz H, Dresselhaus T. ZmES genes encode peptides with structural homology to defensins and are specifically expressed in the female gametophyte of maize. Plant J. 2001;25:103–114. doi: 10.1046/j.0960-7412.2000.00944.x. [DOI] [PubMed] [Google Scholar]
- Diboll AG. Fine structural development of the megagametophyte of Zea mays following fertilization. Am J Bot. 1968;55:787–806. doi: 10.2307/2440968. [DOI] [Google Scholar]
- Dresselhaus T, Márton ML. Micropylar pollen tube guidance and burst: adapted from defense mechanisms? Curr Opin Plant Biol. 2009;12:773–780. doi: 10.1016/j.pbi.2009.09.015. [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Cordts S, Heuer S, Sauter M, Lörtz H, Kranz E. Novel ribosomal genes from maize are differently expressed in the zygotic and somatic cell cycles. Mol Gen Genet. 1999;261:416–427. doi: 10.1007/s004380050983. [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Cordts S, Lörz H. A transcript encoding translation initiation factor eIF-5A is stored in unfertilized egg cells of maize. Plant Mol Biol. 1999;39:1063–1071. doi: 10.1023/A:1006176819213. [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Amien S, Márton M, Strecke A, Brettschneider R, Cordts S. TRANSPARENT LEAF AREA1 encodes a secreted proteolipid required for anther maturation, morphogenesis, and differentiation during leaf development in maize. Plant Cell. 2005;17:730–745. doi: 10.1105/tpc.104.028340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN, Lee D, Christensen CA. Genetic analysis of female gametophyte development and function. Plant Cell. 1998;10:5–17. doi: 10.1105/tpc.10.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimanelli D, Perotti E, Ramirez J, Leblanc O. Timing of the maternal-to-zygotic transition during early seed development in maize. Plant Cell. 2005;17:1061–1072. doi: 10.1105/tpc.104.029819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama T. The synergid cell: attractor and acceptor of the pollen tube for double fertilization. J Plant Res. 2002;115:149–160. doi: 10.1007/s102650200020. [DOI] [PubMed] [Google Scholar]
- Higashiyama T, Yabe S, Sasaki N, Nishimura Y, Miyagishima S, Kuroiwa H, Kuroiwa T. Pollen tube attraction by the synergid cell. Science. 2001;293:1480–1483. doi: 10.1126/science.1062429. [DOI] [PubMed] [Google Scholar]
- Higashiyama T, Kuroiwa H, Kuroiwa T. Pollen-tube guidance: beacons from the female gametophyte. Curr Opin Plant Biol. 2003;6:36–41. doi: 10.1016/S1369-5266(02)00010-9. [DOI] [PubMed] [Google Scholar]
- Huang B-Q, Russell SD. Female germ unit: organization, isolation and function. Int Rev Cytol. 1992;140:233–293. doi: 10.1016/S0074-7696(08)61099-2. [DOI] [Google Scholar]
- Johnston AJ, Meier P, Gheyselinck J, Wuest SE, Federer M, Schlagenhauf E, Becker JD, Grossniklaus U. Genetic subtraction profiling identifies genes essential for Arabidopsis reproduction and reveals interaction between the female gametophyte and the maternal sporophyte. Genome Biol. 2007;8:R204. doi: 10.1186/gb-2007-8-10-r204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenski T, Heilmeier S, Meinhart A, Cramer P. Structure and mechanism of RNA polymerase II CTD phosphatases. Cell. 2004;15:399–407. doi: 10.1016/j.molcel.2004.06.035. [DOI] [PubMed] [Google Scholar]
- Köhler C, Page DR, Gagliardini V, Grossniklaus U. The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat Genet. 2005;37:28–30. doi: 10.1038/ng1495. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê Q, Gutièrrez-Marcos JF, Costa LM, Meyer S, Dickinson HG, Lörz H, Kranz E, Scholten S. Construction and screening of subtracted cDNA libraries from limited populations of plant cells: a comparative analysis of gene expression between maize egg cells and central cell. Plant J. 2005;44:167–178. doi: 10.1111/j.1365-313X.2005.02518.x. [DOI] [PubMed] [Google Scholar]
- Lee TI, Young RA. Transcription of eukaryotic protein—coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- Márton ML, Cordts S, Broadhvest J, Dresselhaus T. Micropylar pollen tube guidance by egg apparatus 1 of maize. Sience. 2005;38:573–576. doi: 10.1126/science.1104954. [DOI] [PubMed] [Google Scholar]
- Meyer S, Scholten S. Equivalent parental contribution to early plant zygotic development. Curr Biol. 2007;17:1686–1691. doi: 10.1016/j.cub.2007.08.046. [DOI] [PubMed] [Google Scholar]
- Mól R, Matthys-Rochon E, Dumas Ch. The kinetics of cytological events during double fertilization in Zea mays L. Plant J. 1994;5:197–206. doi: 10.1046/j.1365-313X.1994.05020197.x. [DOI] [Google Scholar]
- Niedojadło J, Perret-Vivancos C, Kalland KH, Cmarko D, Cremer T, Driel R, Fakan S. Transcribed DNA is preferentially located in the perichromatin region of mammalian cell nuclei. Exp Cell Res. 2011;317:433–444. doi: 10.1016/j.yexcr.2010.10.026. [DOI] [PubMed] [Google Scholar]
- Ning J, Peng XB, Qu LH, Xin HP, Yan TT, Sun M. Differential gene expression in egg cells and zygotes suggests that the transcriptome is restructed before the first zygotic division in tobacco. FEBS Lett. 2006;580:1747–1752. doi: 10.1016/j.febslet.2006.02.028. [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development. 2005;132:603–614. doi: 10.1242/dev.01595. [DOI] [PubMed] [Google Scholar]
- Palancade B, Beiller S, Almouzni G, Bensaude O. Incomplete RNA polymerase II phosphorylation in Xenopus laevis early embryos. J Cell Sci. 2001;114:2483–2489. doi: 10.1242/jcs.114.13.2483. [DOI] [PubMed] [Google Scholar]
- Picton H, Briggs D, Gosden R. The molecular basis of oocyte growth and development. Mol Cell Endocrinol. 1998;145:27–37. doi: 10.1016/S0303-7207(98)00166-X. [DOI] [PubMed] [Google Scholar]
- Pillot M, Baroux C, Vazquez MA, Autran D, Leblanc O, Vielle-Calzada JP, Grossniklaus U, Grimanelli D. Embryo and endosperm inherit distinct chromatin and transcriptional states from the female gametes in Arabidopsis. Plant Cell. 2010;22:307–320. doi: 10.1105/tpc.109.071647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/S0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Ray A. Three’s company: regulatory cross-talk during seed development. Plant Cell. 1997;9:665–667. [Google Scholar]
- Russel SD. Attraction and transport of male gametes for fertilization. Sex Plant Reprod. 1996;9:337–342. doi: 10.1007/BF02441952. [DOI] [Google Scholar]
- Sauter M, Wiegen P, Lörz H, Kranz E. Cell cycle regulatory genes from maize are differentially controlled during fertilization and first embryonic cell division. Sex Plant Reprod. 1998;11:41–48. doi: 10.1007/s004970050119. [DOI] [Google Scholar]
- Scholten S, Lörz H, Kranz E. Paternal mRNA and protein synthesis coincides with male chromatin decondensation in maize zygotes. Plant J. 2002;32:221–231. doi: 10.1046/j.1365-313X.2002.01418.x. [DOI] [PubMed] [Google Scholar]
- Smoliński DJ, Niedojadło J, Noble A, Górska-Brylass A. Additional nucleoli and NOR activity during meiotic prophase I in larch (Larix decidua Mill.) Protoplasma. 2007;232:109–120. doi: 10.1007/s00709-007-0270-y. [DOI] [PubMed] [Google Scholar]
- Sprunck S, Bauman U, Edwards K, Langridge P, Dresselhaus T. The transcript composition of egg cells changes significantly following fertilization in wheat (Triticum aestivum L.) Plant J. 2005;41:660–672. doi: 10.1111/j.1365-313X.2005.02332.x. [DOI] [PubMed] [Google Scholar]
- Steffen JG, Kang IH, Macfarlane J, Drews GN. Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 2007;51:281–292. doi: 10.1111/j.1365-313X.2007.03137.x. [DOI] [PubMed] [Google Scholar]
- Sz Pięciński, Smoliński DJ, Zienkiewicz K, Bednarska E. Changes in poly(A) and TMG snRNA distribution in the embryo sac of Hyacinthus orientalis L. before and after fertilization. Sex Plant Reprod. 2008;21:247–257. doi: 10.1007/s00497-008-0082-z. [DOI] [Google Scholar]
- Telford NA, Watson AJ, Schultz GA. Transition from maternal to embryonic control in early mammalian development: a comparison of several species. Mol Reprod Dev. 1990;26:90–100. doi: 10.1002/mrd.1080260113. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Baskar R, Grossniklaus U. Delayed activation of the paternal genome during seed development. Nature. 2000;404:91–94. doi: 10.1038/35003595. [DOI] [PubMed] [Google Scholar]
- Weijers D, Geldner N, Offringa R, Jürgens G. Seed development: early paternal gene activity in Arabidopsis. Nature. 2001;414:709–710. doi: 10.1038/414709a. [DOI] [PubMed] [Google Scholar]
- Weterings K, Russell SD. Experimental of the fertilization process. Plant Cell. 2004;16:107–118. doi: 10.1105/tpc.016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, Lohr M, Wellmer F, Rahnenführer J, Mering C, Grossniklaus U. Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol. 2010;23:506–512. doi: 10.1016/j.cub.2010.01.051. [DOI] [PubMed] [Google Scholar]
- Yadegari R, Drews GN. Female gametophyte development. Plant Cell. 2004;16:131–144. doi: 10.1105/tpc.018192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Kaur N, Kiriakopolos S, McCormick S. EST generation and analyses towards identifying female gametophyte-specific genes in Zea mays L. Planta. 2006;224:1004–1014. doi: 10.1007/s00425-006-0283-3. [DOI] [PubMed] [Google Scholar]
- Zhao J, Xin H, Qu L, Ning J, Peng X, Yan T, Ma L, Li S, Sun MX. Dynamic changes of transcript profiles after fertilization are associated with de novo transcription and maternal elimination in tobacco zygote, and mark the onset of the maternal-to-zygotic transition. Plant J. 2011;65:131–145. doi: 10.1111/j.1365-313X.2010.04403.x. [DOI] [PubMed] [Google Scholar]
- Zienkiewicz K, Suwińska A, Niedojadło K, Zienkiewicz A, Bednarska E. Nuclear activity of sperm cells during Hyacinthus orientalis L. in vitro pollen tube growth. J Exp Bot. 2011;62:1255–1269. doi: 10.1093/jxb/erq354. [DOI] [PMC free article] [PubMed] [Google Scholar]