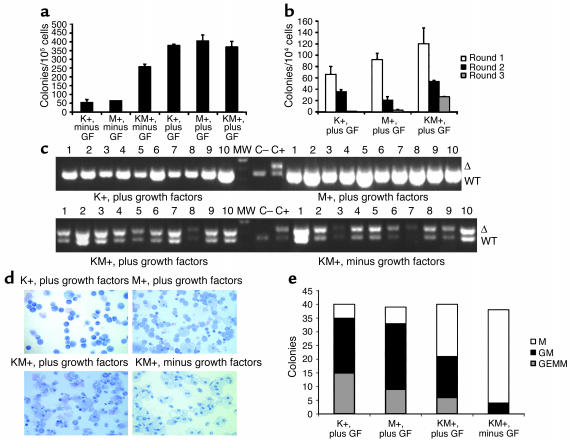
Figure 7.
Methylcellulose cultures with KM+, K+, and M+ bone marrow in the presence and absence of growth factors. (a) Number of colonies generated after methylcellulose culture of 100,000 bone marrow cells in the presence (plus) and absence (minus) of growth factors (GF: SCF, IL-3, and IL-6). Growth factor–independent colony-forming activity of KM+ bone marrow cells was demonstrated in two independent experiments. The values shown are the mean of duplicate cultures from one representative experiment. (b) Number of secondary, tertiary, and quaternary colonies generated in serial methylcellulose cultures using 104 input bone marrow cells in the presence of growth factors. Serial replating assays were performed in two independent experiments. The values shown are the means of duplicate cultures from one representative experiment. (c) PCR for WT and activated (Δ) K-ras alleles demonstrates presence of activated K-ras allele in individual methylcellulose colonies derived from KM+ bone marrow in the presence and absence of growth factors, but not from K+ or M+ colonies. The intensity of the PCR products generated from KM+ growth factor–independent colonies was less robust than that from KM+ growth factor–dependent colonies, corresponding to lesser amounts of input template genomic DNA purified from the smaller individual growth factor–independent colonies. (d) Cytospins (Wright-Giemsa stain) of individual methylcellulose colonies show GM-CFUs derived from K+, M+, and KM+ bone marrow cultured in the presence of growth factors, and M-CFUs from KM+ bone marrow cultured in the absence of growth factors. (e) Quantitation of M-CFU (M), GM-CFU (GM), and GEMM-CFU (GEMM) colonies from K+, M+, and KM+ bone marrow cultured in the presence of growth factors and KM+ bone marrow cultured in the absence of growth factors.