Fig. 5.
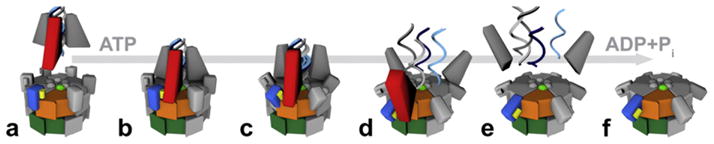
Potential mechanism of NSF. NSF-ATP binds to the C-terminus of α-SNAP (red wedge), which is bound to the coiled coil of the SNARE complex (a and b). Upon ATP hydrolysis the N-domains (blue block) pivot around the D1-domain, away from the center pulling, the α-SNAP and SNARE complex (c and d). This radial force serves to separate the SNAREs and thus disassemble the complex (e and f). The positively charged surface of NSF-N, which is important for SNAP–SNARE binding, is depicted as a yellow bar. The pore residues in NSF-D1 are depicted as green spheres. NSF-D1 and NSF-D2 are depicted as orange and green blocks, respectively.
