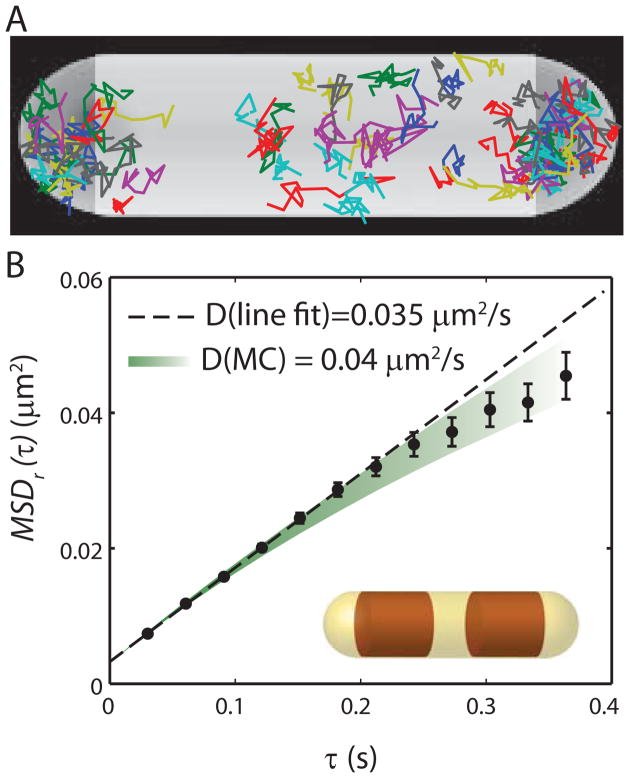
Figure 5.
Ribosome diffusion in untreated cells. (A) Trajectories of single ribosomes (S2-YFP labels) within one cell, plotted within a spherocylinder chosen to match the phase contrast image. (B) Mean-square displacement vs lag time, averaged over 53 single-molecule trajectories from the same cell, each consisting of 13 steps. Error estimates are ±1σof the MSD values from single molecules. Dashed line is a linear fit to first three data points, yielding a diffusion coefficient estimate of D = 0.035 μm2-s−1. Colored swath represents the range of theoretical MSDr(τ) plots (± one standard deviation of the mean) obtained from averaging fifty 13-step Monte Carlo simulated trajectories using D = 0.04 μm2-s−1, the best-fit value as judged by eye. The simulations were run in a spherocylinder within which two truncated cylinders (representing the segregated DNA nucleoid lobes) block ribosome diffusion, as shown in the inset.