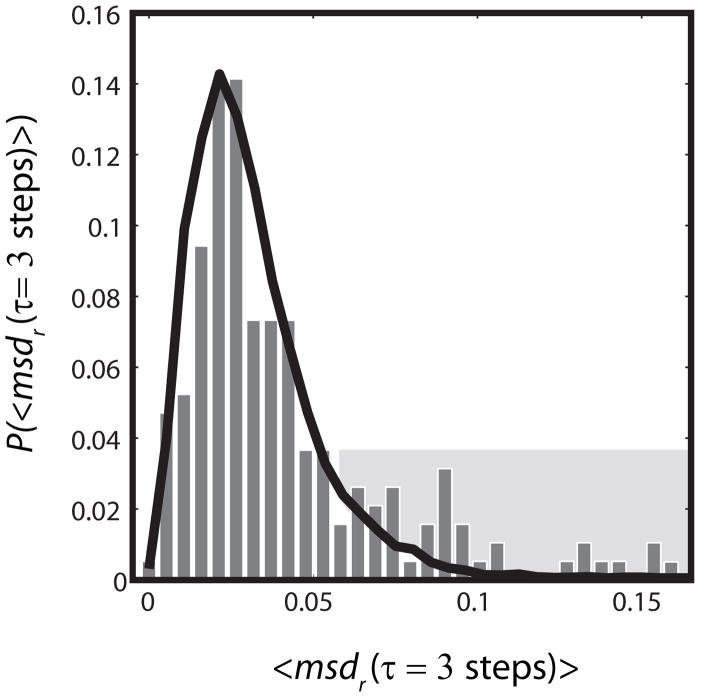
Figure 6.
Heterogeneity of ribosome diffusion. Experimental distribution (bars) of best-estimate single-ribosome diffusion coefficients calculated from the three-step mean-square displacement. Each trajectory of ten steps or longer in a single cell is truncated at 10 steps. For each single-molecule j, msdr,j at lag time τ= 3 steps = 30 ms is calculatedas a running average over all 10 steps. The single-molecule msd is estimated as: msdr,j = <r2(τ=3 steps)>j/4τ. The black line represents the distribution from a simulation with 10,000 trajectories of length 10 step, using a homogeneous diffusion coefficient of 0.04 μm2/s and normalized to match experiment. The geometric model includes two impenetrable cylinders to represent nucleoid lobes, as shown in Fig. 5. The peak and the shaded tail of the experimental distribution are not well fit by the model. See Fig. S8 for a two-component fit to composite data from three cells.