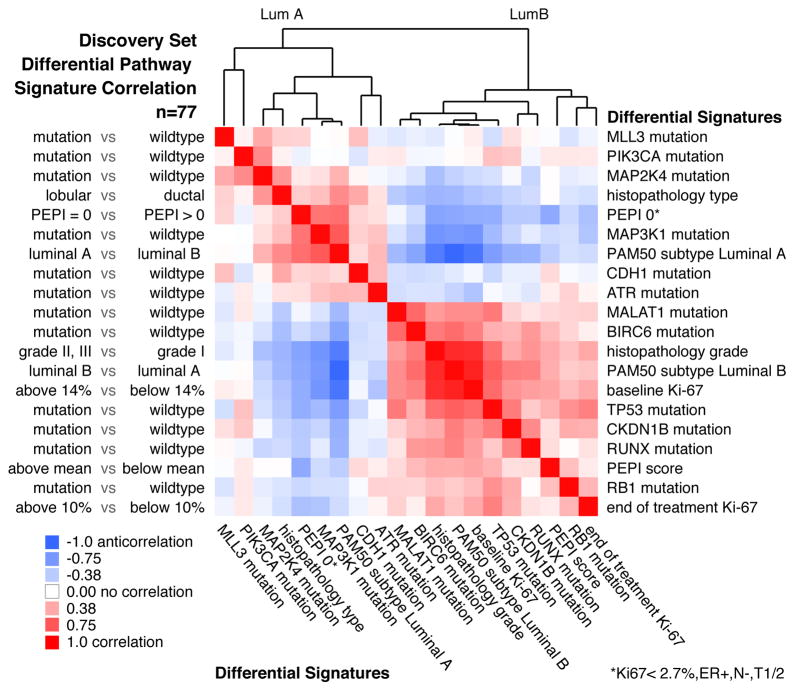
Figure 5. Pathway signatures reveal connections between mutations and clinical outcomes.
PARADIGM-based pathway signatures were derived for tumour feature dichotomies including mutation driven gene signatures (mutant vs. non-mutant), histopathology type (lobular vs. ductal), preoperative endocrine prognostic index (PEPI) score (PEPI=0 favorable vs. PEPI>0 unfavorable), PAM50 Luminal A subtype (LumA vs. LumB) and the reverse (LumB vs. LumA), histopathology grade (grades II&III vs. I), baseline Ki67 levels (>=14% vs. <14%), and end-of-treatment Ki67 levels (>=10% vs. <10%) and overall PEPI score (higher than mean unfavorable vs. lower than mean favorable). Pearson correlations were computed between all pair-wise signatures; positive correlations, red; negative correlations, blue; column features ordered identically as rows. Correlation analysis on the 77 samples in the discovery set is shown.