Figure 8.
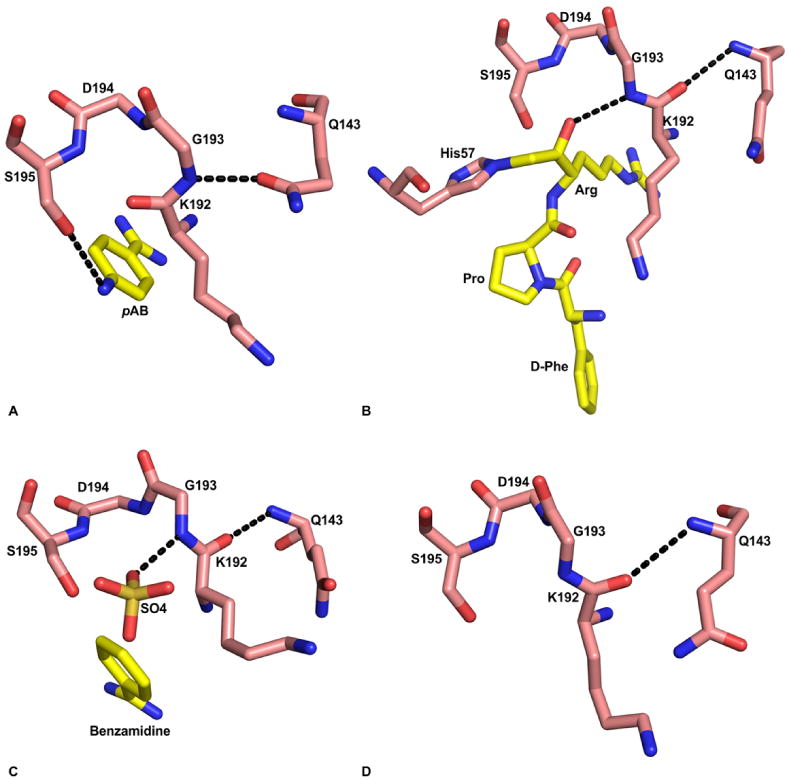
Substrate induced formation of the oxyanion hole in FVIIa protease domain. A) Absence of oxyanion hole in the FVIIa/sTF structure with pAB (PDB id 2A2Q). The hydroxyl group of S195 makes hydrogen bond with the para amino group of pAB. The amide nitrogen of G193 makes hydrogen bond with Odelta of Q143, which prevents formation of the oxyanion hole. The residues 192-195, and Q143 are shown in stick representation. The carbon atoms of protease residues are colored salmon, whereas the carbon atoms in the inhibitor are colored yellow. The ‘N’ and ‘O’ atoms are colored blue and red, respectively. The hydrogen bonds are shown with black dashed lines. B) The fully formed oxyanion hole in FVIIa/sTF with d-FPR (PDB id 2FIR). Note that there is a 180° flip in the peptide bond between K192 and G193 residues as compared to the structure with pAB shown in ‘A’. Now, the carbonyl oxygen of K192 makes hydrogen bond with the backbone amide nitrogen of Q143, and the Odelta of Q143 makes hydrogen bond with R147 (not shown). C) The fully formed oxyanion hole in free FVIIa with benzamidine and sulfate ion in the active site (PDB id 1KLI). The negatively charged sulfate ion in the active site is hydrogen bonded to the amide nitrogen of G193; this interaction induces the formation of oxyanion hole in this structure. D) The K192-G193 peptide bond in standard conformation after removal of the sulfate ion from the FVIIa active site (PDB id 1KLJ). The crystals were obtained under the same condition as that in panel ‘C’; however benzamidine and the sulfate ions were removed by washing the crystal in the buffer lacking benzamidine and sulfate.
