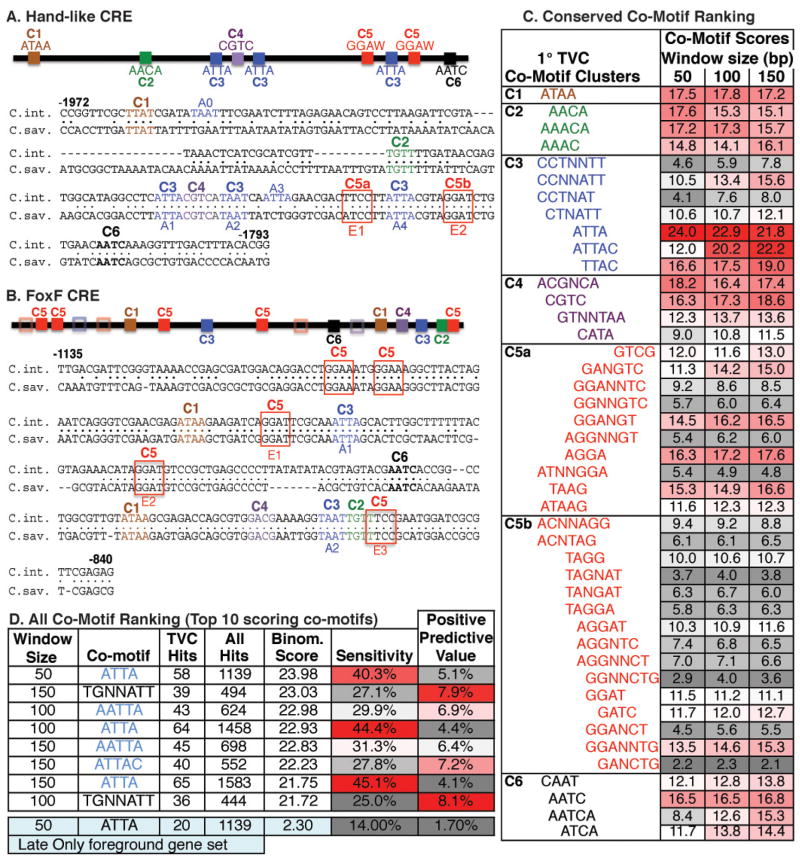
Figure 4. Bio-informatic prediction of Ets Co-motifs in primary TVC regulatory elements.
(A,B) Schematic and sequence alignments of the Hand-like or FoxF CREs showing the distribution of 6 clusters (C1–C6) composed of 47 shared co-motifs. Clusters are grouped by the first appearance of a co-motif in the Hand-like CRE. Hollow boxes indicate partial matches to motif clusters (indicated by color) in the FoxF CRE. ATTA (A0-4) and Ets binding motifs (E1–E3) are also annotated. (C) Co-motif scores for all shared co-motifs grouped by clustering in the Hand-like CRE. Note that some sequences have been reverse-complemented to clarify their alignment within a cluster. Cluster 2 may represent a FoxF binding site involved in maintenance of TVC gene expression. Cluster 4 may represent a CREB binding site (CGTCA, Matsumoto et al., 2007) potentially involved in co-recruitment of a CBP co-activator (Foulds et al., 2004). (D) Data on the top 10 scoring co-motifs from the primary TVC CRE sequence set, along with the ATTA co-motif from the “late only” TVC CRE sequence set.