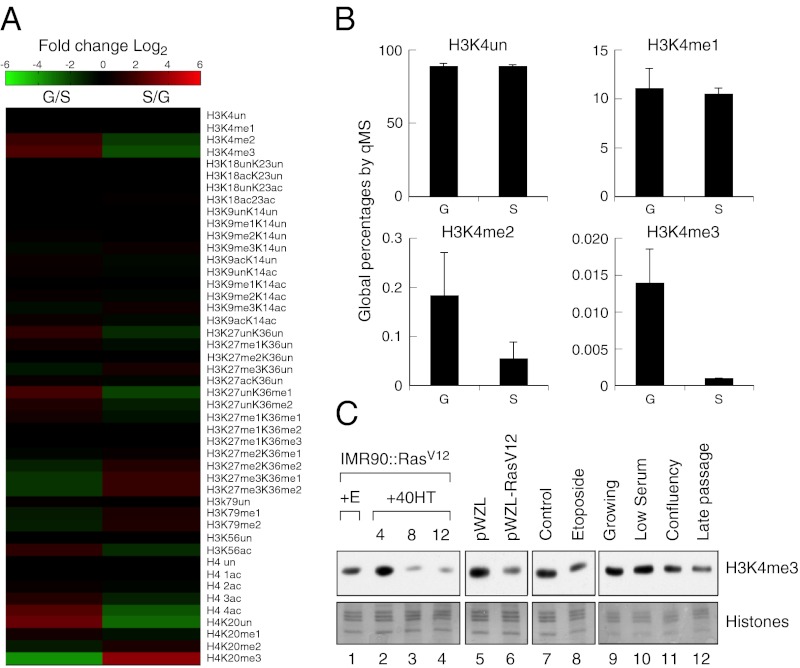
Fig. 1.
Senescence-specific global loss of H3K4 methylation. (A) Heatmap representing the relative levels (log2) of the different histone modifications in growing (G) vs. senescent (S) cells. (B) Histograms representing the global percentages of the different methylation states of H3K4 as determined by qMS. H3K4un is unmodified H3K4. The error bars represent the SD from three technical replicates. (C) Immunoblots using H3K4 methylation specific antibodies showing global loss of H3K4me3 in senescent but not quiescent cells. IMR90 cells treated with ethanol (lane 1) or with 4OHT for the indicated time (lanes 2–4), infected with a vector control (lane 5) or a vector expressing activated Ras (lane 6), treated with DMSO (lane 7) or 50 μM etoposide (lane 8), growing (lane 9), low serum quiescence (lane 10), confluency-induced quiescence (lane 11), and replicative senescence (lane 12). Core histones were used as a loading control.