Abstract
Biomarkers of cancer can indicate the presence of disease and serve as therapeutic targets. Our goal is to develop an optical imaging approach using molecularly targeted contrast agents to assess several centimeters of mucosal surface for mapping expression of multiple biomarkers simultaneously with high spatial resolution. The ability to image biomarker expression level and heterogeneity in vivo would be extremely useful for clinical cancer research, patient selection of personalized medicine, and monitoring therapy. In this proof-of-concept ex vivo study, we examined correlation of neoplasia with two clinically relevant biomarkers: epidermal growth factor receptor (EGFR) and metabolic activity. Two hundred eighty-six unique locations in nine samples of freshly resected oral mucosa were imaged after topically applying optical imaging agents EGF-Alexa 647 (to target EGFR) and 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose (to target metabolic activity). Quantitative features were calculated from resulting fluorescence images and compared with tissue histopathology maps. The EGF-Alexa 647 signal correlated well with EGFR expression as indicated by immunohistochemistry. A classification algorithm for presence of neoplasia based on the signal from both contrast agents resulted in an area under the curve of 0.83. Regions with a posterior probability from 0.80 to 1.00 contained more than 50% neoplasia 99% (84/85) of the time. This study demonstrates a proof-of-concept of how noninvasive optical imaging can be used as a tool to study expression levels of multiple biomarkers and their heterogeneity across a large mucosal surface and how biomarker characteristics correlate with presence of neoplasia. Applications of this approach include predicting regions with the highest likelihood of disease, elucidating the role of biomarker heterogeneity in cancer biology, and identifying patients who will respond to targeted therapy.
Introduction
The molecular changes that accompany the dysplasia-to-carcinoma sequence can serve as diagnostic aids, therapeutic targets, indicators of recurrence, or as surrogate end points in clinical trials of targeted therapeutics [1–4]. For example, in patients with breast cancer, estrogen receptor positivity as indicated by immunohistochemistry (IHC) not only is associated with improved prognosis but also qualifies the patient for targeted hormonal therapy such as tamoxifen [5]. When it was recently discovered that 90% of gastrointestinal stromal tumors have an activating mutation in the KIT (CD117) receptor tyrosine kinase gene, investigators evaluated a KIT inhibitor, imatinib, for treatment and found dramatically improved patient survival [6].
Whereas many promising biomarkers of neoplasia have been identified, only a few are routinely used in clinical decision making. In part, this is due to the lack of tools available to quantitatively assess biomarker expression in vivo at the point-of-care, which limits discovery of the role that biomarkers play in cancer progression. Although hematologic cancers tend to be clonal, solid tumors are frequently extremely heterogeneous. Standard biomarker assessment is currently performed on small biopsy specimens that may not reflect the heterogeneity of a large solid tumor. To achieve the potential of molecular management of neoplasia, there is an important need to better understand how the spatial and temporal changes in biomarker expression correlate with disease progression and to develop methods to noninvasively assess in vivo expression of these biomarkers before, during, and after therapy.
To assess the feasibility of using optical imaging to simultaneously map expression of multiple biomarkers in mucosal surfaces, we performed a proof-of-concept study in freshly resected human surgical specimens from patients with oral cancer. We demonstrate that such maps correlate both with biomarker expression and with histologic diagnosis, and we show that they can be used to predict tissue regions with the highest likelihood of disease.
In the optical molecular imaging approach used in this study, a solution containing a targeted, fluorescent contrast agent is applied topically to a tissue at risk. The use of topical application is an alternate mode of delivering contrast agents compared with intravenous injection that is applicable to both ex vivo and in vivo imaging. It has many benefits, including a decreased required dose and only local exposure, thereby potentially reducing systemic toxicity concerns. Topical application is also associated with shorter incubation times. In the oral cavity, a mouthwash or topical gel formulation could be used to topically apply targeted imaging agents in patients. The tissue is then rinsed and imaged to yield a spatial map of biomarker expression that can be used to guide clinical decision making, aiding in the early detection of neoplasia, staging of disease, selecting and monitoring therapy, detecting recurrence, and predicting the likelihood of progression. Although the study reported here was conducted using freshly resected oral tissue, the concept could be translated to live patient oral cavity imaging once regulatory requirements are fulfilled. Such an approach using topical application of a fluorescent imaging agent in vivo to detect colonic neoplasms was recently demonstrated [7].
Optical molecular imaging has potential advantages over the two most widely used techniques to assess biomarker expression levels: IHC and reverse transcription-polymerase chain reaction. Most frequently, IHC is used to assess protein expression in vitro in histologic specimens. The limitations of IHC include the need to remove tissue for assessment, difficulty in quantifying biomarker expression levels, and challenges associated with examining multiple biomarkers per slide, unless the proteins of interest lie in distinct compartments within the cell [8]. Information from multiple biomarkers frequently outperforms information from a single marker alone [5]. Breast cancer presents a good example of this in clinical practice, where estrogen receptor, progesterone receptor, and HER2/neu expression are all routinely assessed for prognosis and selection of therapy. Conversely, reverse transcription-polymerase chain reaction and other high-throughput methods to altered mRNA expression associated with neoplasia can be used to simultaneously assess thousands of potential biomarkers. However, gene expression profiling requires tissue removal, and it is difficult to assess information about spatial distribution of biomarker changes. It is also not clear that changes in gene expression lead to changes in protein expression.
Optical molecular imaging using targeted contrast agents, however, is ideally suited to achieve the goal of spatial and temporal assessment of biomarker expression in vivo. Optical molecular imaging can be used to noninvasively image the distribution of multiple biomarkers across a large mucosal surface in vivo. Optical molecular imaging has been used to image eight or more biomarkers in distinct spectral bands with high spatial resolution and may serve as an important tool in better understanding the role of biomarkers in the disease process [9].
The goal of this study was to explore the role of optical imaging with multiple topically applied targeted contrast agents to study the heterogeneity of biomarker expression and its correlation with the presence of neoplasia across a large mucosal surface area. The oral cavity was chosen as an organ site because of its accessibility to optical imaging. Optically active contrast agents targeting two clinically relevant biomarkers, the epidermal growth factor receptor (EGFR) and metabolic activity, were applied to the tissue surface, and the resulting optical molecular images were compared with the gold standard of histopathology.
EGFR is one of the most commonly studied biomarkers in cancers of the oral cavity because it is widely overexpressed in oral squamous cell carcinoma and dysplasia [10]. Although the reported percentage of patients with overexpression of EGFR varies between studies, a review from Lippman et al. [11] indicates that 80% to 100% of patients with premalignant or malignant oral lesions have high EGFR expression [11]. A recent study from Taoudi Benchekroun et al. [12] found that 71% of oral premalignant lesions display high EGFR expression levels, which correlated with a greater risk of developing oral cancer. EGFR expression level also increases with the progression of disease [13–16]. The prevalence of EGFR expression makes it a potentially useful target to improve detection and guide treatment; in fact, the EGFR inhibitor, cetuximab, is approved by the US Food and Drug Administration (FDA)for head and neck cancer therapy.
Another clinical biomarker for oral cancer is increased metabolic activity. Metabolism is elevated in dysplasia and cancer due to rapid cell growth and division; consequently, glucose transporters (GLUTs) are often overexpressed or operate at a higher activity level. In a study by Kunkel et al., 100% (40/40) of oral cancers expressed GLUT-1 by IHC, and Mellanen et al. found that every tumor sample they examined expressed either GLUT-1 or GLUT-3 mRNA [17,18]. Ayala et al. [19] found GLUT-1 overexpression in 50% of their oral cancer specimens, Tian et al. [20] observed that 73.7% of samples had moderate to strong GLUT-1 expression, and Ohba et al. [21] discovered that overexpression of GLUT-1 in the invasion front is associated with the tumor depth and prognosis.
Materials and Methods
Preparation of Fluorescent Imaging Agents
To assess EGFR expression, the natural ligand of EGFR, the EGF, was conjugated to a fluorescent dye. Purified human-derived recombinant EGF peptide (Calbiochem, San Diego, CA) was conjugated to Alexa Fluor 647-carboxylic ester (Invitrogen, Carlsbad, CA). Excess free dye was removed by size exclusion chromatography followed by dialysis. The resulting EGF-Alexa 647 imaging agent was diluted to a final concentration of 25 µg/ml in sterile 1x phosphate-buffered saline (PBS) with 10% dimethylsulfoxide as a permeation enhancer. Specificity studies in both cells and resected human tissue have been previously performed and published with EGF-Alexa 647, demonstrating the ability of this agent to specifically target EGFR in cell culture and in oral mucosa [22].
A fluorescently labeled glucose molecule was used to assess metabolic activity. 2-(N-(7-nitrobenz-2-oxa-1,3 -diazol-4-yl)amino)-2-deoxyglucose (2-NBDG) is a commercially available optical glucose analog (Invitrogen, Carlsbad, CA). 2-NBDG was diluted to a concentration of 0.16 mM in sterile 1x PBS. Multiple studies have used 2-NBDG as a metabolic activity indictor [23–25]. Tumor cells in culture have been shown to take up approximately five times the amount of 2-NBDG compared with nonmalignant cells, and competition assays with free glucose decreased the amount of 2-NBDG that was taken up [23]. A recent study by Sheth et al. [24] demonstrated the comparability of 2-NBDG to FDG-PET imaging in several preclinical examples, and Nitin et al. [25] demonstrated the potential use of 2-NBDG for detecting neoplasia in clinical specimens, in which dysplasia and cancer displayed a two-to five-fold increase in fluorescence intensity compared with normal tissue.
Application of Fluorescent Imaging Agents to Clinical Samples
To evaluate the ability of 2-NBDG and EGF-Alexa 647 to identify neoplasia in human oral tissue, freshly resected clinical specimens were obtained from patients undergoing surgery at MD Anderson Cancer Center. These patients gave written informed consent to participate in the study, which was approved by the Institutional Review Boards at both MD Anderson Cancer Center and Rice University. Neoplastic tissue was obtained immediately after surgical resection and taken to a laboratory for contrast agent application and subsequent imaging.
To assess autofluorescence before the application of any fluorescent contrast agents, the tissue was imaged with a multispectral digital microscope (MDM), a wide-field imaging system that has been described in detail previously [26]. Briefly, the MDM is a dental microscope that has been modified with fluorescence excitation and emission filters and a mercury-argon light source; it is capable of imaging in both reflectance and fluorescence modes. It has a large field of view (∼5 x 7 cm) and can achieve a lateral spatial resolution up to 0.016 mm. Preincubation images of the tissue were taken using standard white light setting and the filter configurations for 2-NBDG (excitation = 475 nm, emission = 550 nm) and EGF-Alexa 647 (excitation = 650, emission = 670 nm) to allow later subtraction of tissue autofluorescence from post-incubation images.
After autofluorescence imaging, 2-NBDG solution was topically applied to the epithelial surface of the resected oral tissue and placed in an incubator at 37°C for 20 minutes. After incubation with 2-NBDG, the tissue was briefly rinsed with sterile 1x PBS to remove any excess unbound fluorescent contrast agent. The tissue was again imaged with the MDM using the filter configurations for 2-NBDG and EGF-Alexa 647. Next, EGF-Alexa 647 in 10% dimethylsulfoxide was topically applied to the epithelial surface of the tissue and incubated for 20 minutes at 37°C. The tissue was briefly rinsed with sterile 1x PBS and imaged using appropriate filter settings on the MDM.
Once imaging was complete, the entire tissue was returned to the Pathology Department of MD Anderson Cancer Center. The tissue was processed following standard procedures: the tissue was thinly sliced anterior to posterior and placed into cassettes for hematoxylin and eosin (H&E) staining. Locations of pathology cuts were documented, and the portion of the tissue that was placed into each cassette was recorded to allow for future correlation to pathology. Neither 2-NBDG nor EGF-Alexa 647 affects histopathology processing or staining.
Creation of Histopathology Maps
To compare the results of wide-field imaging with the gold standard of histopathology, a two-dimensional color-coded pathology map was created for each clinical specimen. To create this map, all the H&E slides for each specimen were reviewed by an expert head and neck pathologist blinded to the optical images (M.W.). The diagnosis across the entire epithelium was recorded for each slide. This expert diagnosis from the slide was assumed to represent the diagnosis for the entire thickness of the thin piece of tissue that was placed in the corresponding cassette. The diagnose of normal epithelium; mild, moderate, and severe dysplasia; and cancer were all assigned a unique color code. On the basis of detailed notes of where the tissue was sliced and of the diagnosis across the epithelium for each piece of tissue, the color-coded histopathology map was created as an overlay on top of the white light image of the clinical specimen. The resulting two-dimensional histopathology map was used as a gold standard to compare with the wide-field fluorescence images. Creation of a histopathology map is not standard practice in pathology, and to create an accurate map, the tissue had to have a fairly flat surface, slicing had to be strictly monitored and documented, and a detailed pathology reading across the entire surface had to be obtained and recorded. All the tissue samples used in this study met these criteria.
Immunohistochemistry
The Histology Core Laboratory at MD Anderson Cancer Center performed immunohistochemical staining for EGFR on multiple specimens to confirm that optical molecular imaging results correlated with the gold standard of IHC for the presence of EGFR. Immunohistochemical analysis was performed using the automated BOND MAX immunohistochemical stainer by Vision Biosystems (Norwell, MA) using the standard clinically used antibody and protocol for EGFR (clone 31G7 mouse antihuman; 1:50; Zymed, South San Francisco, CA). An expert head and neck pathologist reviewed these slides to grade the intensity of staining across the entire epithelial surface. The scale used for assessment of intensity of EGFR immunostaining was from 0 to 3. Score 0 indicated no staining or nonspecific staining, score 1 was used for weak staining of more than 10% of cells, score 2 indicated moderate staining of more than 10% of cells, and score 3 indicated strong and complete staining of more than 10% of cells [8]. The level of immunostaining was compared to fluorescence intensity from the corresponding portion of tissue to further validate the correlation between IHC for EGFR and EGF-Alexa 647 fluorescence.
IHC for GLUT expression was not compared with fluorescence imaging results with 2-NBDG, because while IHC can reveal GLUT expression level, it cannot assess GLUT activity level. Previous studies have not found a significant correlation between overexpression of GLUTs and standardized uptake values of FDG in FDG-PET imaging, a widely accepted clinical method of analyzing metabolic activity [20,27].
Quantitative Analysis of Fluorescence Images
Postincubation fluorescence images of both 2-NBDG and EGF-Alexa 647 were analyzed using Matlab R2010b software (The MathWorks, Natick, MA) to identify trends in optical molecular images that correlate with diagnosis. All images before and after incubation were aligned using a Matlab image registration algorithm. The fluorescence images were converted to grayscale, and background autofluorescence was subtracted. For each clinical sample, multiple square regions of interest 50 x 50 pixels in size were selected, using the histopathology map as a guide to ensure that each region selected contained a uniform diagnosis and that regions did not overlap. Image features were calculated and compared with feature values from regions of normal tissue to determine whether these characteristics were altered with the presence of neoplasia. These features included normalized mean intensity, standard deviation, skewness, kurtosis, and the coefficient of variation within each region. Normalization of mean intensity was performed per patient; the mean of all histologically normal regions of interest for a patient was used to normalize every region of interest selected for that patient. In addition, a Matlab edge detection algorithm using the Canny method was applied to the bulk fluorescence image to detect where sharp gradients in intensity occurred because this may also serve as a measure of heterogeneity of biomarker expression. The length of this edge line divided by the number of pixels in the region was calculated for all selected regions of interest.
Development of a Classification Algorithm
To discriminate normal from neoplastic regions of interest based on these quantitative feature values, a classification algorithm was developed using linear discriminant analysis. The algorithm used all of the 50 x 50 pixel regions for both training and testing. Features were selected sequentially to identify which were most useful in distinguishing neoplastic tissue from normal tissue. Only three features were chosen to avoid overtraining of the algorithm. Sensitivity, specificity, and area under the receiver operator characteristic curve (AUC) were calculated.
Application of Classification Algorithm to a New Data Set to Predict Regions with the Highest Likelihood of Disease
We examined the ability of this algorithm to identify regions of interest with the highest likelihood of disease, information that could be useful for early detection and diagnosis, planning surgical resection, monitoring treatment response, or screening for recurrence. A 50 x 50 pixel grid was used to divide the original fluorescence images from all the patients into new regions of interest. The classification algorithm described previously was applied to these regions to generate posterior probabilities (a value between 0 and 1) for all the new regions. The practical application of this strategy would be to help a clinician objectively identify areas with the highest likelihood of disease. The regions with posterior probabilities from 0.80 to 1.00 were identified and compared with histopathology map information at that site. Regions with posterior probabilities from 0.60 to 0.79 were also identified as regions likely to be neoplastic, and this prediction was compared with the histopathology gold standard to assess performance of the algorithm. Only regions that contained epithelium throughout the entire 50 x 50 pixel region were evaluated; regions overlapping the edge of the tissue were not considered.
Results
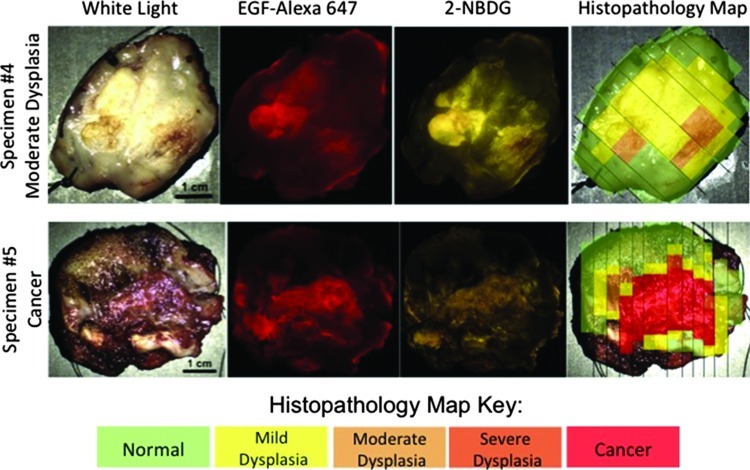
Nine clinical specimens from different patients were imaged as described; a histopathology map was created for each specimen. Figure 1 shows images from two representative clinical specimens: one with mild to moderate dysplasia and one with cancer. Qualitative comparison of the fluorescence images and the gold standard histopathology maps of each specimen demonstrates that, for both imaging agents, there is greater fluorescence intensity in areas of neoplasia, including areas of invasive carcinoma as well as in areas of mild to moderate dysplasia. Note that due to the manner in which the imaging agent was applied, topical application at the edges of the specimen resulted in some imaging agent coming in contact with the deep tissue. The imaging agent is not as easily rinsed from the deep tissue, and so a small edge effect can be observed; higher fluorescence intensity is seen around the edges of the tissue.
Figure 1.
Images from two representative specimens. From left to right, images include a white light photograph, fluorescence after incubation with EGF-Alexa 647 to show EGFR expression, fluorescence afterincubation with 2-NBDG to show metabolic activity, and the correspondinghistopathologymap. The specimen shown in the top row hada worst diagnosis of moderate dysplasia; the specimenin the bottom row had a worst diagnosis of cancer. Scale bars, 1 cm. The color scale on the bottom shows the key for the histopathology maps.
IHC to Confirm EGFR Expression
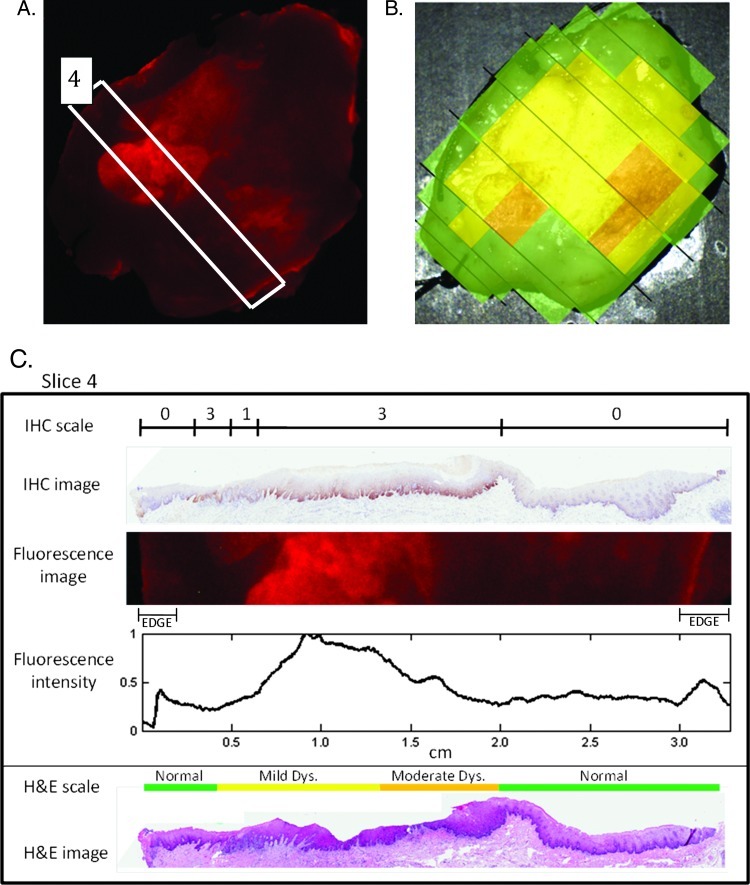
IHC staining for EGFR was performed on multiple specimens to confirm previously published findings validating the specificity of EGF-Alexa 647 for EGFR. Figure 2 shows results from one representative patient specimen. Figure 2A shows the wide-field EGF-Alexa 647 fluorescence image for this specimen; a white rectangle indicates the slice of tissue for which IHC was performed. Figure 2B shows the corresponding histopathology map for this specimen. Figure 2C shows the immunostained slide prepared from this slice along with the IHC scale, indicating the pathologist's grading of degree of staining for EGFR. Below the IHC image is the corresponding portion of the fluorescence image for comparison. The fluorescence intensity was summed for each column in the fluorescence image and normalized to the maximum value; results are shown on the fluorescence intensity graph in Figure 2C. Figure 2C also shows the H&E-stained slide and H&E scale (corresponding to the histopathology map) for reference. Regions of high fluorescence intensity correspond well to regions of elevated IHC staining across this tissue slice, verifying the specificity of EGF-Alexa 647. The edge effect mentioned earlier is the cause of the slightly higher fluorescence at the edges of the tissue that do not correlate with IHC results.
Figure 2.
A representative set of images to demonstrate the correlation between EGF-Alexa 647 fluorescence intensity and IHC for EGFR. (A) Wide-field fluorescence image of specimen following application of EGF-Alexa 647 with tissue slice selected for IHC outlined in white. (B) Histopathology map of specimen. (C) IHC staining of selected slice as graded by a pathologist is indicated on a scale of 0 to 3 along with the original IHC image. The fluorescence image from the corresponding portion of tissue is also shown, and fluorescence intensity is graphed across the slice on a scale normalized to the maximum value. The original H&E slide and corresponding histologic diagnosis are also shown for reference.
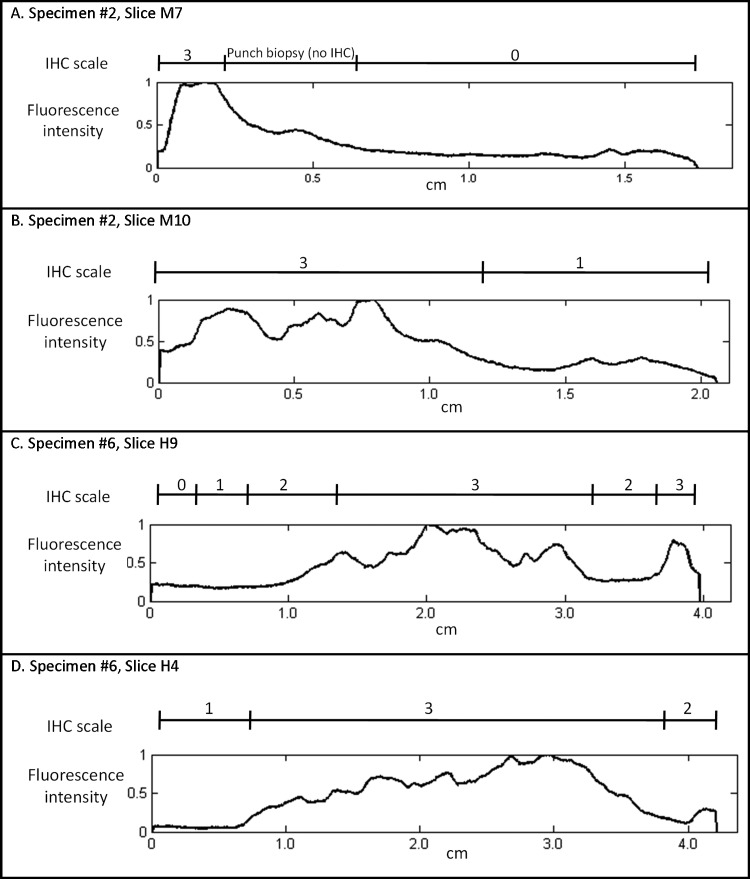
The same type of analysis was performed for four patients to ensure consistency of results, although in this case, the edges were cropped. Figure 3 compares the intensity of IHC staining and the graph of normalized fluorescence intensity for two additional specimens, with two different slices shown from each. IHC grading and fluorescence intensity correlate well for all the slides investigated.
Figure 3.
Intensity of IHC staining and corresponding fluorescence intensity graphs for two additional specimens with two slices shown from each: (A) specimen 2, slice M7; (B) specimen 2, slice M10; (C) specimen 6, slice H9; and (D) specimen 6, slice H4.
Quantitative Analysis of Fluorescence Images
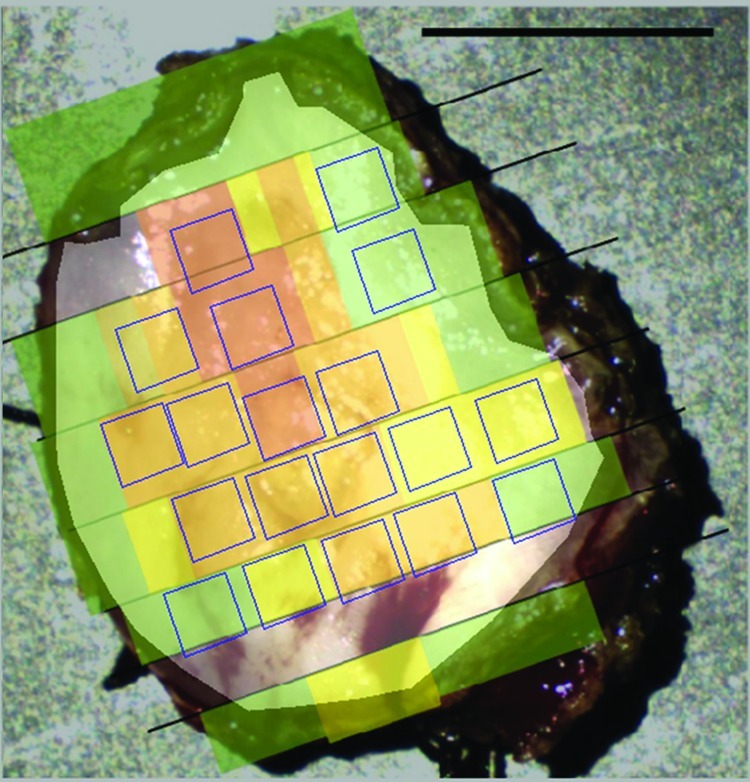
Figure 4 shows a representative sample with the outlines of the selected 50 x 50 pixel regions of interest. This specimen contains four normal regions, three regions of mild dysplasia, nine regions of moderate dysplasia, and three regions of severe dysplasia. The regions are oriented parallel to the pathology slices to maximize the number of regions that could be selected per sample. Table 1 details the number of regions that were selected per sample by diagnostic category. Overall, 286 total regions were selected from the nine specimens and were categorized as follows: 87 normal regions, 57 regions with mild dysplasia, 26 regions with moderate dysplasia, 20 regions containing severe dysplasia, and 95 regions with cancer.
Figure 4.
Representative example of how 50 x 50-pixel regions of interest were selected. This sample contains four normal regions, three regions of mild dysplasia, nine regions of moderate dysplasia, and three regions of severe dysplasia. Quantitative features were calculated from each region of interest. Scale bar, 1 cm.
Table 1.
Number of Analyzed Regions by Patient and Diagnosis.
| Specimen No. | Normal | Mild Dysplasia | Moderate Dysplasia | Severe Dysplasia | Cancer | Total |
| 1 | 4 | 3 | 9 | 3 | 0 | 19 |
| 2 | 18 | 1 | 1 | 0 | 12 | 32 |
| 3 | 14 | 3 | 0 | 15 | 4 | 36 |
| 4 | 12 | 33 | 8 | 0 | 0 | 53 |
| 5 | 9 | 6 | 3 | 2 | 17 | 37 |
| 6 | 9 | 3 | 3 | 0 | 23 | 38 |
| 7 | 3 | 2 | 1 | 0 | 2 | 8 |
| 8 | 6 | 1 | 1 | 0 | 23 | 31 |
| 9 | 12 | 5 | 0 | 0 | 15 | 32 |
| Total | 87 | 57 | 26 | 20 | 96 | 286 |
Development of a Classification Algorithm
For each region, image features including average intensity, standard deviation, skewness, kurtosis, maximum value, and coefficient of variation were calculated from postincubation fluorescence images. In addition, the length of the edge line detected using the Matlab edge function was calculated for each region using fluorescence images. These image features were used to train and test a classification algorithm on the full set of 286 regions; a histologic diagnosis of normal epithelium was considered normal, and mild, moderate, and severe dysplasia and cancer were considered neoplastic. Table 2 indicates the classification performance of the top 6 highest-performing single features, ranked by AUC. Of the six top-performing features, four are calculated from the EGF-Alexa 647 image and two are calculated from the 2-NBDG image. Normalized mean intensity of the EGF-Alexa 647 signal was the highest performing single feature with an AUC of 0.80.
Table 2.
Performance of Single Features and Combination of Features in a Classification Algorithm.
| Feature | AUC of Feature |
| EGF normalized mean intensity | 0.80 |
| EGF entropy | 0.76 |
| EGF standard deviation | 0.73 |
| 2-NBDG normalized mean intensity | 0.70 |
| EGF maximum value | 0.70 |
| 2-NBDG maximum value | 0.68 |
| Three-feature algorithm | 0.83 |
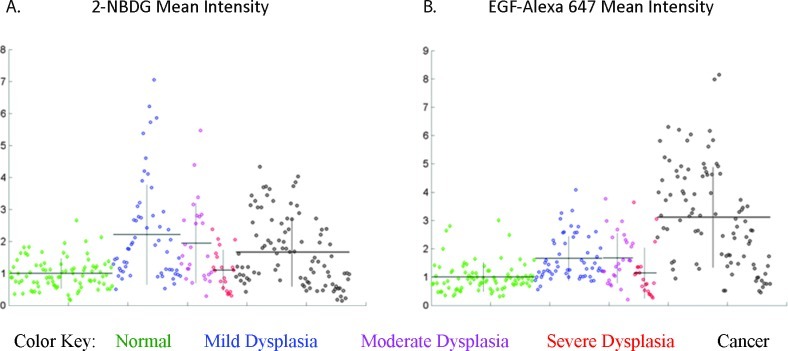
Figure 5 shows scatterplots of two selected features of interest, with the y axis indicating feature value and the x axis sorted by pathologic diagnosis. Each of the 286 points in the graph indicates a single region of interest. Figure 5A shows normalized mean intensity of the 2-NBDG signal and Figure 5B shows normalized mean intensity of the EGF-Alexa 647 signal. Regions are grouped by pathologic diagnosis with green indicating normal epithelium, blue indicating mild dysplasia, pink indicating moderate dysplasia, red indicating severe dysplasia, and black indicating cancer. Horizontal black lines indicate the mean feature value and vertical black lines indicate ±1 standard deviation for each pathologic category. Figure 5 indicates that the 2-NBDG signal intensity, and therefore metabolic activity, is elevated earlier in the progression of neoplasia, whereas EGFR expression is elevated later in this progression.
Figure 5.
Scatterplots showing the distribution of feature values within the set of 286 regions; each point represents a single region. Regions are grouped by pathologic diagnosis with green indicating normal epithelium, blue indicating mild dysplasia, pink indicating moderate dysplasia, red indicating severe dysplasia, and black indicating cancer. Horizontal black lines indicate the mean feature value and vertical black lines indicate ±1 standard deviation for each pathologic category. The scatterplots show the following features: (A) mean intensity of the 2-NBDG signal and (B) mean intensity of the EGF-Alexa 647 signal.
Ultimately, three unbiased features were selected for use in the classification algorithm: (1) normalized mean intensity of the EGF-Alexa 647 signal, (2) coefficient of variation from the 2-NBDG signal, and (3) length of the detected edge based on 2-NBDG. The features were selected sequentially in that order for improving performance of the algorithm; the normalized mean intensity of the EGF-Alexa 647 signal was the single best feature, and each of the subsequent features improved algorithm performance. Addition of a fourth feature did not significantly improve performance. Table 2 also indicates the AUC of the three-feature algorithm developed from this combination of quantitative features.
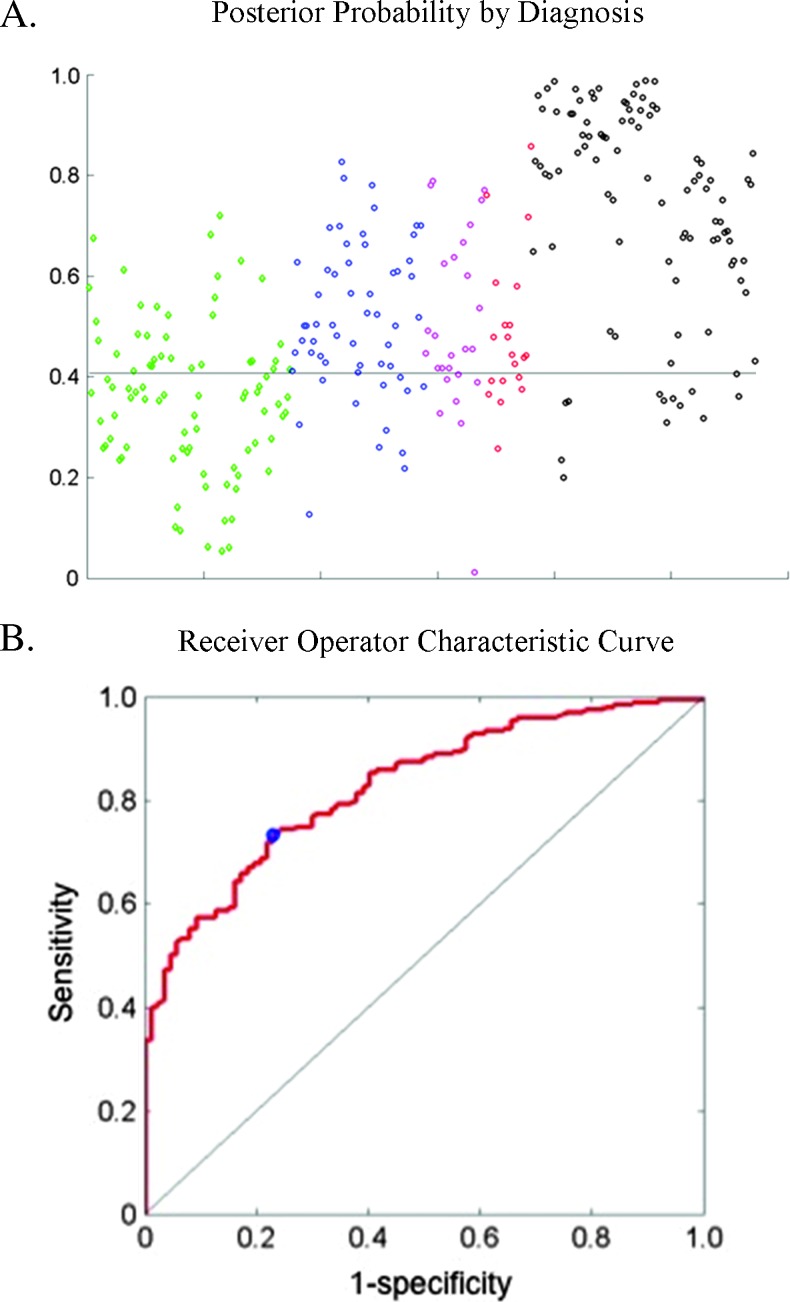
Figure 6A shows the resulting scatterplot of posterior probabilities grouped by histopathologic diagnosis. The horizontal line at 0.41 indicates the cutoff point that gives the best sensitivity and specificity of the algorithm. Any posterior probability above this value would be considered neoplastic by the classification algorithm, and any posterior probability below this value would be considered normal. This cutoff gives a sensitivity of 73% and a specificity of 77%. Figure 6B shows the receiver operator characteristic curve of the algorithm. The AUC is 0.83, and the cutoff point yielding the highest sensitivity and specificity is marked with a blue circle.
Figure 6.
(A) Scatterplot of posterior probability by diagnosis using the algorithm. The horizontal line indicates the optimum cutoff at 0.41. The color key is the same as in Figure 5. (B) Receiver operator characteristic curve using a three-feature classificat ion algorithm. The operating point at the optimum cutoff is shown with a blue circle and results in a sensitivity of 73%, a specificity of 77%, and an AUC of 0.83.
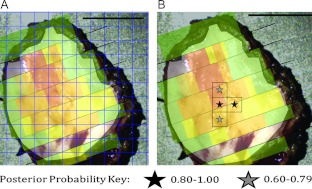
Figure 7A displays the 50 x 50-pixel grid that was used to divide the image from a representative sample into new regions. Whereas the originally evaluated regions as shown in Figure 4 were selected to contain only one diagnostic category and were oriented parallel to the pathology slices, the new evaluation grid is oriented parallel to the edges of the image and each region may contain multiple diagnostic categories. Figure 7B indicates the regions that were identified as having the highest posterior probability. Black stars indicate a posterior probability between 0.80 and 1.00 and gray stars indicate a posterior probability between 0.60 and 0.79. In this example, two adjacent regions have the highest level of posterior probability. According to the histopathology map, one of these regions contains mostly severe dysplasia, and the other contains mostly moderate dysplasia. Two regions with the second highest level of posterior probability were also identified, and these are also adjacent to one of the highest-level regions. Of these two regions, marked with a gray star, one contains mostly severe dysplasia and the other contains moderate dysplasia throughout. No part of any of these identified regions in this specimen contains normal epithelium.
Figure 7.
(A) Superimposed grid to divide each specimen into a new set of regions of interest. Using the previously developed classification algorithm, posterior probabilities were calculated for each of these new regions of interest. (B) On the basis of two levels of posterior probability, regions identified as most likely to contain neoplasia were marked with black (posterior probability = 0.80–1.00) or gray (posterior probability = 0.60–0.79) stars. Scale bars, 1 cm.
This method was applied to all nine specimens; results are shown in Table 3. Of the nine specimens, seven specimens contained at least one region that had a posterior probability between 0.80 and 1.00, for a total of 85 regions. Of these identified regions, 99% (84/85) contained more than 50% neoplasia. The remaining two specimens that did not have any regions identified within the highest-level category (nos. 3 and 7) had multiple regions with posterior probabilities between 0.60 and 0.79. Among the nine specimens, a total of 85 different regions were identified as having a posterior probability from 0.60 to 0.79. Of these regions, 95% (81/85) contained more than 50% neoplasia.
Table 3.
Use of Posterior Probability to Identify Regions with the Highest Likelihood of Disease.
| Specimen No. | Regions with Posterior Probability of 0.80–1.00 | Regions with Posterior Probability of 0.60–0.79 |
| 1 | 100% (2/2) | 100% (2/2) |
| 2 | 93% (13/14) | 80% (4/5) |
| 3 | N/A | 50% (1/2) |
| 4 | 100% (1/1) | 100% (15/15) |
| 5 | 100% (25/25) | 80% (8/10) |
| 6 | 100% (31/31) | 100% (8/8) |
| 7 | N/A | 100% (3/3) |
| 8 | 100% (11/11) | 100% (24/24) |
| 9 | 100% (1/1) | 100% (16/16) |
| Total | 99% (84/85) | 95% (81/85) |
Values are presented as percentage of regions containing more than 50% neoplasia.
N/A indicates not applicable.
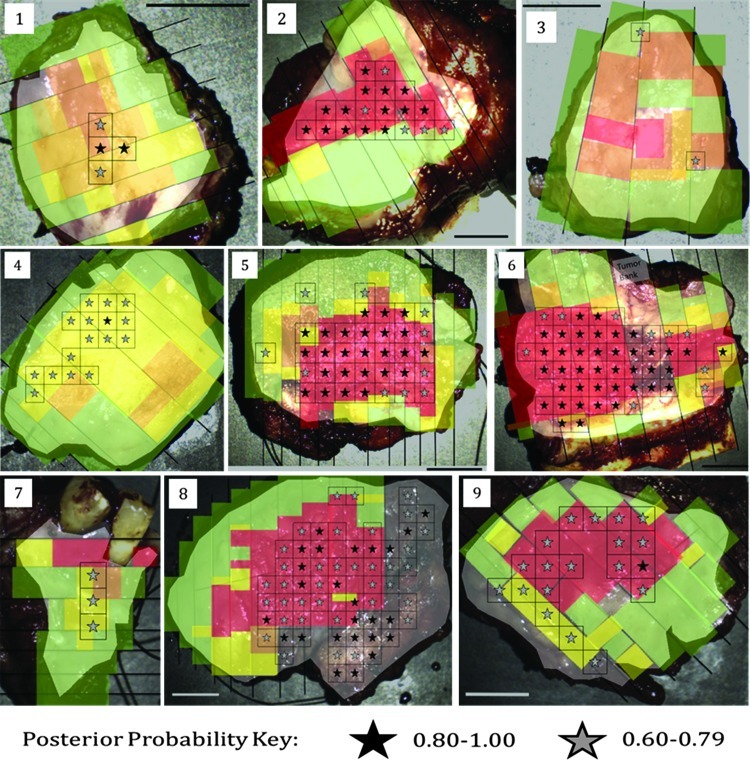
Figure 8 shows for all the clinical samples the regions determined to have the highest posterior probability overlaid onto the corresponding histopathology maps. The black stars indicate the regions with posterior probability from 0.80 to 1.00, and the gray stars indicate the regions with posterior probability from 0.60 to 0.79. A few of the regions identified do not have corresponding gold standard histopathology information for the entire area and so were not included in the analysis shown in Table 3, but are shown in Figure 8.
Figure 8.
All clinical specimens; regions identified by the algorithm as most likely to contain neoplasia are marked by stars. Black stars indicate the highest level of posterior probability (0.80–1.00) and gray stars indicate the next highest level of posterior probability (0.60–0.79). Regions predicted to contain neoplasia have excellent agreement with the criterion standard of histopathology. Scale bars, 1 cm.
This strategy has excellent agreement with histopathology, and although it does not identify all abnormal tissue, it identifies areas with the highest likelihood of disease in all samples.
Discussion
The optical imaging approach described in this study uses molecularly targeted contrast agents to simultaneously assess two clinically relevant biomarkers in oral cancer: EGFR expression and metabolic activity. The benefit of this approach is that expression levels of multiple biomarkers can be assessed quantitatively over a large field of view and with high spatial resolution; this type of information cannot be obtained through the current practice of IHC. In this study, we used ex vivo imaging to evaluate whether topical application of multiple imaging agents could be potentially useful to predict the presence of neoplastic disease in the oral cavity. A potential advantage of studying optical imaging approaches in freshly resected human tissue as an alternative to in vivo assessments in animal models is that the pattern of biomarker expression and the potential barriers to topical administration of imaging agents will likely be more physiologically relevant to the intended final goal of in vivo imaging of patients with oral cancer [28].
The optical molecular imaging approach demonstrated here ex vivo can be translated to in vivo oral cavity imaging. The imaging system used in this study has been used to measure oral autofluorescence in vivo in many patients [26,29,30]. The targeted imaging agents can be formulated into a mouthwash solution or topical gel that could be topically applied to the oral mucosal surface, potentially decreasing the dose required in comparison with intravenous injection and reducing consequent systemic toxicity concerns. Imaging could be performed shortly after application. Steps remaining before this technique can be translated to in vivo use in patients include safety testing to support an Investigational New Drug application for topical use of the imaging agents. This would allow clinical testing to assess the utility of the technique in patients with oral dysplasia or cancer. This approach could also be extended to include other biomarkers, if spectrally distinct imaging agents that specifically target the biomarker can be developed.
Analysis of fluorescence images in Figure 6 revealed features that differ in images of normal and neoplastic tissue. Whereas using the classification algorithm to simply classify a site as either normal or neoplastic resulted in a sensitivity of 73% and a specificity of 77%, the plot of posterior probability versus diagnosis suggests a more useful way in which to apply the image features. When these image features were used to identify regions with the highest likelihood of disease, the selected areas contained histologically diagnosed neoplasia 99% of the time. The advantages of the quantitative features used for classification are that they provide objective metrics that reflect the amount of the molecular target present and the heterogeneity of the target expression. These features can be calculated rapidly and easily. The objective assessment to accurately predict the presence of neoplasia has many clinical applications, including initial detection and diagnosis, selection of an appropriate biopsy site, monitoring treatment response, and monitoring patients for recurrence. This application is consistent with research recommended in 2010 in the Journal of the American Dental Association, which called for research projects to “identify factors that would increase clinicians' confidence and competence in identification and management of potentially malignant lesions or early-stage malignancies, including the provision of surgical biopsies” [31].
Another potential use of a targeted optical imaging strategy could be in understanding the role of heterogeneity of biomarker expression in selection of therapy and predicting treatment response. Quantitative IHC studies reveal that the factor of overexpression of EGFR ranges from approximately 1.7- to 3-fold increase in dysplasia and cancer [13,14,32]. A similar level of overexpression has also been observed in targeted molecular imaging studies, and the results of this study are comparable [22,33]. EGFR expression is already a target in the FDA-approved monoclonal antibody therapy cetuximab, which is often used to treat recurrence of oral cancer. Other EGFR inhibitors (gefitinib, erlotinib, panatumumab, and lapatinib) have been approved for cancer treatment in other organ sites and are currently in clinical trials for head and neck cancer indications [34]. Whereas in vitro data suggest a link between EGFR expression level and cytotoxicity of EGFR-targeted therapy, no definitive clinical correlation has yet been found [35–37]. As seen in the fluorescence images and IHC data shown here, EGFR expression is heterogeneous throughout a lesion. The ability to assess biomarker expression in vivo over a large sample using optical molecular imaging may elucidate the causes of this discrepancy and aid in the selection of patients who could benefit from EGFR-targeted therapy. For patients who respond, the benefits of EGFR-targeted therapy include the prevention of nodal metastases by blocking EGFR and potentially improved survival when combined with standard chemotherapy [38,39]. As new EGFR-targeted therapies (such as nimotuzumab, zalutulumab, bevacizumab, and vandetanib) are developed and tested clinically for head and neck cancer indications, predicting which patients may benefit from a particular therapy will also aid in FDA approval [40,41].
EGFR expression also has applications in other clinical settings. EGFR may help distinguish between leukoplakia with or without dysplasia because oral leukoplakia with underlying dysplasia has increased expression of EGFR, which may be useful as an early marker of malignancy [15]. Moreover, nonmalignant leukoplakias were shown not to have elevated EGFR expression [42]. Another clinical application of EGFR expression is in the identification of patients who would benefit from hyperfractionated accelerated radiotherapy [43,44]. In pretreatment biopsies assessed for EGFR, patients with high EGFR expression had improved survival with hyperfractionated accelerated radiotherapy compared with a standard radiotherapy schedule. Identifying EGFR expression across the entire surface of the lesion may improve identification of patients who could benefit from this type of treatment. Finally, quantification of EGFR expression is difficult with current IHC methods, which may be one reason for conflicting findings in studies examining the prognostic value of EGFR in head and neck cancer [8]. Quantitative evaluation of optical molecular imaging of EGFR may help standardize measurement of EGFR expression level.
Metabolic activity as indicated by FDG-PET is currently used extensively in head and neck cancer for tumor staging, monitoring treatment response, and detecting residual or recurrent disease [45,46]. FDG-PET has a high sensitivity and moderate specificity for detecting recurrent disease at the primary tumor site, but because of its inherently low spatial resolution, it is unable to detect lesions smaller than approximately 1 cm in size [46]. FDG-PET is significantly better than clinical assessment and conventional imaging to monitor treatment response and has also been used as a marker of erlotinib response [47,48]. Monitoring metabolic activity with FDG-PET can also identify residual disease and detect recurrence [46,47,49–51]. Although monitoring metabolic activity optically with 2-NBDG cannot accomplish any of the deep tissue applications of FDG-PET such as imaging of nodal metastases, it can be used to assess metabolic activity in the epithelium as shown in this study, a strategy that may be useful to detect residual or recurrent disease at the primary tumor site. The advantage of using 2-NBDG over FDG-PET is that optical imaging provides superior spatial and temporal resolution, does not expose the patient to ionizing radiation, and is far less expensive than PET imaging equipment and maintenance of cyclotron facilities.
Optical molecular imaging as applied in this study has some limitations, however. Although topical application of imaging agents may reduce dose and toxicity concerns, the agents are delivered only in the epithelium, preventing detection of submucosal disease. Another concern may arise from the idea of applying the EGF, a mitogen, to a potentially neoplastic area. However, studies have demonstrated that pretreatment of oral cancer lesions with EGF actually improves the cytotoxicity of cisplatin, the most commonly used chemotherapy for head and neck cancer [52,53]. Inflammation in the oral cavity is a common confounder for detection of neoplasia, and inflammatory cells may also take up 2-NBDG, resulting in potential false-positives from high metabolic activity of lymphocytes present in the oral tissue due to benign conditions. However, targeting GLUTs is still very clinically relevant as evidenced by the widespread use of FDG-PET imaging. Having an orthogonal target such as EGFR is useful to prevent the occasional false-positive from 2-NBDG signal due to inflammation because EGFR is not elevated as a result of inflammation. Finally, not all patients will have overexpression of EGFR, so that low fluorescence signal after application of EGF-Alexa 647 may not rule out the possibility of the presence of neoplasia.
Being able to obtain information from both agents in combination may help avoid the pitfalls of each agent alone. As seen from the images in Figure 1 and the quantitative results in Figure 5, although the general spatial pattern of staining with both agents appears similar with elevation across regions of neoplasia, there are important differences. 2-NBDG signal is most elevated in regions with histologic diagnosis of dysplasia, and EGFR signal is most elevated in regions with histologic diagnosis of cancer. The changes in 2-NBDG signal over progression of disease is related first to rapid growth and metabolism of cells during dysplasia, followed by necrosis and ulceration that occurs once a large tumor develops. EGFR expression increases over disease progression and is at its highest once cancer develops. 2-NBDG and EGFR therefore provide complementary information.
Biomarkers have been studied for their potential use in detecting neoplasia, staging, selecting and monitoring therapy, detection of recurrence, and prognostic value, but the full picture of biomarker expression across an entire lesion is necessary to make accurate conclusions about the roles these biomarkers play in the disease process. Optical molecular imaging as demonstrated in this study of EGFR expression and metabolic activity in oral mucosa can reveal biomarker heterogeneity across a lesion and may serve as a research tool to understand cancer biology, develop new targeted therapies, or aid in the clinical management of cancer patients. With the proper imaging agents, this approach could be extended to monitor additional biomarkers in other types of epithelial neoplasia as well and may eventually result in improved patient survival by allowing clinical care to be tailored to each patient's biomarker expression profile.
Acknowledgments
The authors would like to thank Darren Roblyer, Tim Muldoon, Richard Schwarz, and Dongsuk Shin for their help in collecting patient data. We thank Vivian Mack for providing help with cell cultures.
Footnotes
This material is also based on the work supported by the National Institutes of Health under grant no. 5T32 GM008362. The authors thank the National Cancer Institute for funding support (BRP R01CA103830). The authors declare no conflict of interest.
References
- 1.Lippman SM, Hong WK. Molecular markers of the risk of oral cancer. N Engl J Med. 2001;344:1323–1326. doi: 10.1056/NEJM200104263441710. [DOI] [PubMed] [Google Scholar]
- 2.Shah NG, Trivedi TI, Tankshali RA, Goswami JA, Shah JS, Jetly DH, Kobawala TP, Patel KC, Shukla SN, Shah PM, et al. Molecular alterations in oral carcinogenesis: significant risk predictors in malignant transformation and tumor progression. Int J Biol Markers. 2007;22:132–143. doi: 10.1177/172460080702200207. [DOI] [PubMed] [Google Scholar]
- 3.Mashberg A, Samit AM. Early detection, diagnosis, and management of oral and oropharyngeal cancer. CA Cancer J Clin. 1989;39:67–88. doi: 10.3322/canjclin.39.2.67. [DOI] [PubMed] [Google Scholar]
- 4.Braakhuis BJ, Leemans CR, Brakenhoff RH. A genetic progression model of oral cancer: current evidence and clinical implications. J Oral Pathol Med. 2004;33:317–322. doi: 10.1111/j.1600-0714.2004.00225.x. [DOI] [PubMed] [Google Scholar]
- 5.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 6.Hornick JL, Fletcher CD. The role of KIT in the management of patients with gastrointestinal stromal tumors. Hum Pathol. 2007;38:679–687. doi: 10.1016/j.humpath.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Hsiung PL, Hardy J, Friedland S, Soetikno R, Du CB, Wu AP, Sahbaie P, Crawford JM, Lowe AW, Contag CH, et al. Detection of colonic dysplasia in vivo using a targeted heptapeptide and confocal microendoscopy. Nat Med. 2008;14:454–458. doi: 10.1038/nm1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atkins D, Reiffen KA, Tegtmeier CL, Winther H, Bonato MS, Storkel S. Immunohistochemical detection of EGFR in paraffin-embedded tumor tissues: variation in staining intensity due to choice of fixative and storage time of tissue sections. J Histochem Cytochem. 2004;52:893–901. doi: 10.1369/jhc.3A6195.2004. [DOI] [PubMed] [Google Scholar]
- 9.Themelis G, Yoo JS, Ntziachristos V. Multispectral imaging using multiple-bandpass filters. Opt Lett. 2008;33:1023–1025. doi: 10.1364/ol.33.001023. [DOI] [PubMed] [Google Scholar]
- 10.Mydlarz WK, Hennessey PT, Califano JA. Advances and perspectives in the molecular diagnosis of head and neck cancer. Expert Opin Med Diagn. 2010;4:53–65. doi: 10.1517/17530050903338068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lippman SM, Sudbo J, Hong WK. Oral cancer prevention and the evolution of molecular-targeted drug development. J Clin Oncol. 2005;23:346–356. doi: 10.1200/JCO.2005.09.128. [DOI] [PubMed] [Google Scholar]
- 12.Taoudi Benchekroun M, Saintigny P, Thomas SM, El-Naggar AK, Papadimitrakopoulou V, Ren H, Lang W, Fan YH, Huang J, Feng L, et al. Epidermal growth factor receptor expression and gene copy number in the risk of oral cancer. Cancer Prev Res (Phila) 2010;3:800–809. doi: 10.1158/1940-6207.CAPR-09-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin DM, Ro JY, Hong WK, Hittelman WN. Dysregulation of epidermal growth factor receptor expression in premalignant lesions during head and neck tumorigenesis. Cancer Res. 1994;54:3153–3159. [PubMed] [Google Scholar]
- 14.Rubin Grandis J, Melhem MF, Barnes EL, Tweardy DJ. Quantitative immunohistochemical analysis of transforming growth factor-α and epidermal growth factor receptor in patients with squamous cell carcinoma of the head and neck. Cancer. 1996;78:1284–1292. doi: 10.1002/(SICI)1097-0142(19960915)78:6<1284::AID-CNCR17>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 15.Srinivasan M, Jewell SD. Evaluation of TGF-α and EGFR expression in oral leukoplakia and oral submucous fibrosis by quantitative immunohistochemistry. Oncology. 2001;61:284–292. doi: 10.1159/000055335. [DOI] [PubMed] [Google Scholar]
- 16.Kannan S, Chandran GJ, Balaram P, Chidambaram S, Nair MK. Potential biological markers for the staging of tumor progression in oral mucosa: a multivariate analysis. Int J Biol Markers. 1996;11:67–76. doi: 10.1177/172460089601100202. [DOI] [PubMed] [Google Scholar]
- 17.Mellanen P, Minn H, Grenman R, Harkonen P. Expression of glucose transporters in head-and-neck tumors. Int J Cancer. 1994;56:622–629. doi: 10.1002/ijc.2910560503. [DOI] [PubMed] [Google Scholar]
- 18.Kunkel M, Moergel M, Stockinger M, Jeong JH, Fritz G, Lehr HA, Whiteside TL. Overexpression of GLUT-1 is associated with resistance to radiotherapy and adverse prognosis in squamous cell carcinoma of the oral cavity. Oral Oncol. 2007;43:796–803. doi: 10.1016/j.oraloncology.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Ayala FR, Rocha RM, Carvalho KC, Carvalho AL, da Cunha IW, Lourenco SV, Soares FA. GLUT1 and GLUT3 as potential prognostic markers for oral squamous cell carcinoma. Molecules. 2010;15:2374–2387. doi: 10.3390/molecules15042374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian M, Zhang H, Nakasone Y, Mogi K, Endo K. Expression of Glut-1 and Glut-3 in untreated oral squamous cell carcinoma compared with FDG accumulation in a PET study. Eur J Nucl Med Mol Imaging. 2004;31:5–12. doi: 10.1007/s00259-003-1316-9. [DOI] [PubMed] [Google Scholar]
- 21.Ohba S, Fujii H, Ito S, Fujimaki M, Matsumoto F, Furukawa M, Yokoyama J, Kusunoki T, Ikeda K, Hino O. Overexpression of GLUT-1 in the invasion front is associated with depth of oral squamous cell carcinoma and prognosis. J Oral Pathol Med. 2010;39:74–78. doi: 10.1111/j.1600-0714.2009.00814.x. [DOI] [PubMed] [Google Scholar]
- 22.Nitin N, Rosbach KJ, El-Naggar A, Williams M, Gillenwater A, Richards-Kortum RR. Optical molecular imaging of epidermal growth factor receptor expression to improve detection of oral neoplasia. Neoplasia. 2009;11:542–551. doi: 10.1593/neo.09188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Neil RG, Wu L, Mullani N. Uptake of a fluorescent deoxyglucose analog (2-NBDG) in tumor cells. Mol Imaging Biol. 2005;7:388–392. doi: 10.1007/s11307-005-0011-6. [DOI] [PubMed] [Google Scholar]
- 24.Sheth RA, Josephson L, Mahmood U. Evaluation and clinically relevant applications of a fluorescent imaging analog to fluorodeoxyglucose positron emission tomography. J Biomed Opt. 2009;14:064014. doi: 10.1117/1.3259364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nitin N, Carlson AL, Muldoon T, El-Naggar AK, Gillenwater A, Richards-Kortum R. Molecular imaging of glucose uptake in oral neoplasia following topical application of fluorescently labeled deoxy-glucose. Int J Cancer. 2009;124:2634–2642. doi: 10.1002/ijc.24222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roblyer D, Richards-Kortum R, Sokolov K, El-Naggar AK, Williams MD, Kurachi C, Gillenwater AM. Multispectral optical imaging device for in vivo detection of oral neoplasia. J Biomed Opt. 2008;13:024019. doi: 10.1117/1.2904658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li SJ, Guo W, Ren GX, Huang G, Chen T, Song SL. Expression of Glut-1 in primary and recurrent head and neck squamous cell carcinomas, and compared with 2-[18F]fluoro-2-deoxy-d-glucose accumulation in positron emission tomography. Br J Oral Maxillofac Surg. 2008;46:180–186. doi: 10.1016/j.bjoms.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Shanks N, Greek R, Greek J. Are animal models predictive for humans? Philos Ethics Humanit Med. 2009;4:2. doi: 10.1186/1747-5341-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roblyer D, Kurachi C, Stepanek V, Williams MD, El-Naggar AK, Lee JJ, Gillenwater AM, Richards-Kortum R. Objective detection and delineation of oral neoplasia using autofluorescence imaging. Cancer Prev Res. 2009;2:423–431. doi: 10.1158/1940-6207.CAPR-08-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roblyer D, Kurachi C, Stepanek V, Schwarz RA, Williams MD, El-Naggar AK, Lee JJ, Gillenwater AM, Richards-Kortum R. Comparison of multispectral wide-field optical imaging modalities to maximize image contrast for objective discrimination of oral neoplasia. J Biomed Opt. 2010;15:066017. doi: 10.1117/1.3516593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rethman MP, Carpenter W, Cohen EE, Epstein J, Evans CA, Flaitz CM, Graham FJ, Hujoel PP, Kalmar JR, Koch WM, et al. Evidence-based clinical recommendations regarding screening for oral squamous cell carcinomas. J Am Dent Assoc. 2010;141:509–520. doi: 10.14219/jada.archive.2010.0223. [DOI] [PubMed] [Google Scholar]
- 32.Rubin Grandis J, Tweardy DJ, Melhem MF. Asynchronous modulation of transforming growth factor a and epidermal growth factor receptor protein expression in progression of premalignant lesions to head and neck squamous cell carcinoma. Clin Cancer Res. 1998;4:13–20. [PubMed] [Google Scholar]
- 33.Carlson AL, Gillenwater AM, Williams MD, El-Naggar AK, Richards-Kortum RR. Confocal microscopy and molecular-specific optical contrast agents for the detection of oral neoplasia. Technol Cancer Res Treat. 2007;6:361–374. doi: 10.1177/153303460700600501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Modjtahedi H, Essapen S. Epidermal growth factor receptor inhibitors in cancer treatment: advances, challenges and opportunities. Anticancer Drugs. 2009;20:851–855. doi: 10.1097/CAD.0b013e3283330590. [DOI] [PubMed] [Google Scholar]
- 35.Rogers SJ, Box C, Chambers P, Barbachano Y, Nutting CM, Rhys-Evans P, Workman P, Harrington KJ, Eccles SA. Determinants of response to epidermal growth factor receptor tyrosine kinase inhibition in squamous cell carcinoma of the head and neck. J Pathol. 2009;218:122–130. doi: 10.1002/path.2515. [DOI] [PubMed] [Google Scholar]
- 36.Hickinson DM, Marshall GB, Beran GJ, Varella-Garcia M, Mills EA, South MC, Cassidy AM, Acheson KL, McWalter G, McCormack RM, et al. Identification of biomarkers in human head and neck tumor cell lines that predict for in vitro sensitivity to gefitinib. Clin Transl Sci. 2009;2:183–192. doi: 10.1111/j.1752-8062.2009.00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharafinski ME, Ferris RL, Ferrone S, Grandis JR. Epidermal growth factor receptor targeted therapy of squamous cell carcinoma of the head and neck. Head Neck. 2010;32:1412–1421. doi: 10.1002/hed.21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamakawa H, Nakashiro K, Sumida T, Shintani S, Myers JN, Takes RP, Rinaldo A, Ferlito A. Basic evidence of molecular targeted therapy for oral cancer and salivary gland cancer. Head Neck. 2008;30:800–809. doi: 10.1002/hed.20830. [DOI] [PubMed] [Google Scholar]
- 39.Kondo N, Tsukuda M, Ishiguro Y, Kimura M, Fujita K, Sakakibara A, Takahashi H, Toth G, Matsuda H. Antitumor effects of lapatinib (GW572016), a dual inhibitor of EGFR and HER-2, in combination with cisplatin or paclitaxel on head and neck squamous cell carcinoma. Oncol Rep. 2010;23:957–963. doi: 10.3892/or_00000720. [DOI] [PubMed] [Google Scholar]
- 40.Basavaraj C, Sierra P, Shivu J, Melarkode R, Montero E, Nair P. Nimotuzumab with chemoradiation confers a survival advantage in treatment-naive head and neck tumors over expressing EGFR. Cancer Biol Ther. 2010;10:673–681. doi: 10.4161/cbt.10.7.12793. [DOI] [PubMed] [Google Scholar]
- 41.Williams MD. Integration of biomarkers including molecular targeted therapies in head and neck cancer. Head Neck Pathol. 2010;4:62–69. doi: 10.1007/s12105-010-0166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prado SM, Cedrun JL, Rey RL, Villaamil VM, Garcia AA, Ayerbes MV, Aparicio LA. Evaluation of COX-2, EGFR, and p53 as biomarkers of non-dysplastic oral leukoplakias. Exp Mol Pathol. 2010;89:197–203. doi: 10.1016/j.yexmp.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Bentzen SM, Atasoy BM, Daley FM, Dische S, Richman PI, Saunders MI, Trott KR, Wilson GD. Epidermal growth factor receptor expression in pretreatment biopsies from head and neck squamous cell carcinoma as a predictive factor for a benefit from accelerated radiation therapy in a randomized controlled trial. J Clin Oncol. 2005;23:5560–5567. doi: 10.1200/JCO.2005.06.411. [DOI] [PubMed] [Google Scholar]
- 44.Suwinski R, Jaworska M, Nikiel B, Grzegorz W, Bankowska-Wozniak M, Wojciech M, Krzysztof S, Dariusz L. Predicting the effect of accelerated fractionation in postoperative radiotherapy for head and neck cancer based on molecular marker profiles: data from a randomized clinical trial. Int J Radiat Oncol Biol Phys. 2010;77:438–446. doi: 10.1016/j.ijrobp.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 45.Scarfone C, Lavely WC, Cmelak AJ, Delbeke D, Martin WH, Billheimer D, Hallahan DE. Prospective feasibility trial of radiotherapy target definition for head and neck cancer using 3-dimensional PET and CT imaging. J Nucl Med. 2004;45:543–552. [PubMed] [Google Scholar]
- 46.Al-Ibraheem A, Buck A, Krause BJ, Scheidhauer K, Schwaiger M. Clinical applications of FDG PET and PET/CT in head and neck cancer. J Oncol. 2009;2009:208725. doi: 10.1155/2009/208725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin RC, Fulham M, Shannon KF, Hughes C, Gao K, Milross C, Tin MM, Jackson M, Clifford A, Boyer MJ, et al. Accuracy of positron emission tomography in the evaluation of patients treated with chemoradiotherapy for mucosal head and neck cancer. Head Neck. 2009;31:244–250. doi: 10.1002/hed.20962. [DOI] [PubMed] [Google Scholar]
- 48.Vergez S, Delord JP, Thomas F, Rochaix P, Caselles O, Filleron T, Brillouet S, Canal P, Courbon F, Allal BC. Preclinical and clinical evidence that deoxy-2-[18F]fluoro-d-glucose positron emission tomography with computed tomography is a reliable tool for the detection of early molecular responses to erlotinib in head and neck cancer. Clin Cancer Res. 2010;16:4434–4445. doi: 10.1158/1078-0432.CCR-09-2795. [DOI] [PubMed] [Google Scholar]
- 49.Ito K, Yokoyama J, Kubota K, Morooka M, Shiibashi M, Matsuda H. 18F-FDG versus 11C-choline PET/CT for the imaging of advanced head and neck cancer after combined intra-arterial chemotherapy and radiotherapy: the time period during which PET/CT can reliably detect non-recurrence. Eur J Nucl Med Mol Imaging. 2010;37:1318–1327. doi: 10.1007/s00259-010-1400-x. [DOI] [PubMed] [Google Scholar]
- 50.de Bree R, van der Putten L, Brouwer J, Castelijns JA, Hoekstra OS, Leemans CR. Detection of locoregional recurrent head and neck cancer after (chemo)radiotherapy using modern imaging. Oral Oncol. 2009;45:386–393. doi: 10.1016/j.oraloncology.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 51.Zundel MT, Michel MA, Schultz CJ, Maheshwari M, Wong SJ, Campbell BH, Massey BL, Blumin J, Wilson JF, Wang D. Comparison of physical examination and fluorodeoxyglucose positron emission tomography/computed tomography 4–6 months after radiotherapy to assess residual head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2011;81:e825–e832. doi: 10.1016/j.ijrobp.2010.11.072. [DOI] [PubMed] [Google Scholar]
- 52.Ahsan A, Hiniker SM, Ramanand SG, Nyati S, Hegde A, Helman A, Menawat R, Bhojani MS, Lawrence TS, Nyati MK. Role of epidermal growth factor receptor degradation in cisplatin-induced cytotoxicity in head and neck cancer. Cancer Res. 2010;70:2862–2869. doi: 10.1158/0008-5472.CAN-09-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nouri AM, Thompson C, Cannell H, Symes M, Purkiss S, Amirghofran Z. Profile of epidermal growth factor receptor (EGFR) expression in human malignancies: effects of exposure to EGF and its biological influence on established human tumour cell lines. Int J Mol Med. 2000;6:495–500. doi: 10.3892/ijmm.6.4.495. [DOI] [PubMed] [Google Scholar]