Figure 1.
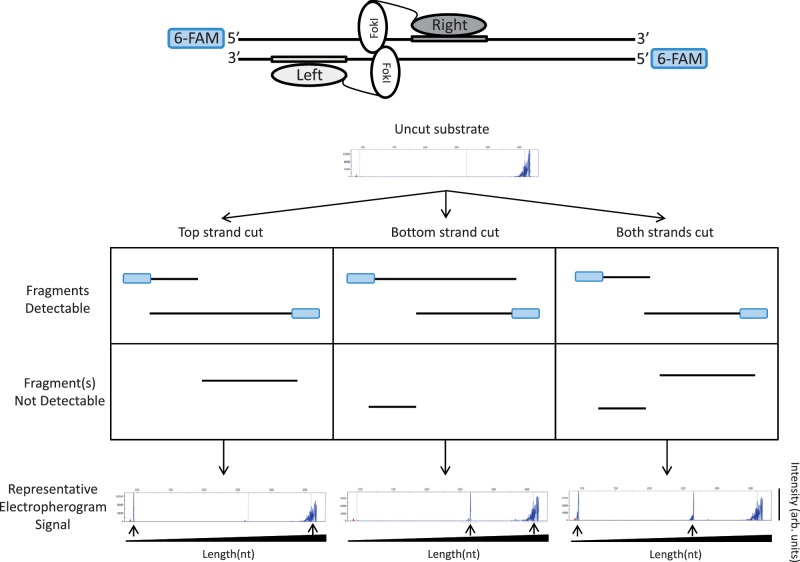
A qualitative in vitro assay to detect cleavage and nicking by ZFNs and ZFNickases. An asymmetrically positioned full ZFN target site is placed within a DNA fragment that has been labeled on both its 5′-ends with 6-FAM fluorescent dye (depicted in blue). Only 6-FAM labeled strands will be detected in the denaturing capillary electrophoresis assay. In the example shown, the ZFN target site is positioned toward the left end of the DNA fragment. In this configuration, if nicking of the top strand occurs, this results in the generation of one short and one full-length 6-FAM labeled product. If nicking of the bottom strand occurs, this results in the generation of one medium-length and one full-length 6-FAM labeled product. Cleavage of both strands results in the generation of short and medium-length products. Sample electropherograms are shown with arbitrary intensity units on the y-axis and DNA strand length on the x-axis. DNA strands expected from nicking or cleavage reactions are designated by black arrows. Note that full-length DNA strands due to incomplete enzyme reactions may be present in addition to the expected products.