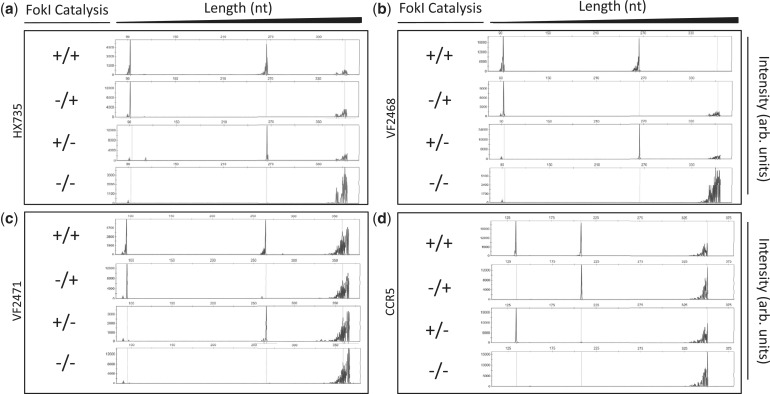
Figure 2.
Site-specific nicking of DNA in vitro by ZFNickases. Substrates labeled with 6-FAM fluorescent dye harboring (a) HX735, (b) VF2468, (c) VF2471 or (d) CCR5 binding sites were incubated with active Left/active Right (+/+), inactive Left/active Right (−/+), active Left/inactive Right (+/−) and inactive Left/inactive Right (−/−) ZFN monomers. Cleavage products were subjected to denaturing capillary electrophoresis. Axes are arbitrary intensity units (y-axis) and DNA strand length (x-axis). The y-axis is differentially scaled for each plot, whereas the x-axis is scaled uniformly for all plots. Representative electropherograms are shown, but all experiments were performed in triplicate (data not shown). Note that the HX735, VF2468 and VF2471 targets were cloned into the pCP5 vector that results in asymmetric placement left of center within the substrate similar to the configuration depicted in Figure 1. However, the CCR5 target is cloned into pBAC-lacZ, which results in binding site placement right of center relative to the substrate; when the top strand is cleaved in this configuration, the fragment generated is longer than when the bottom strand is cleaved.