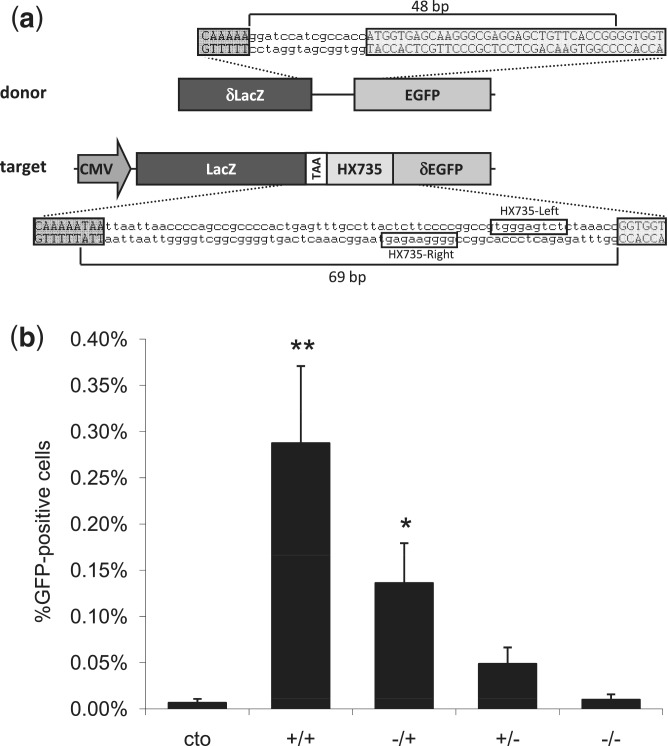
Figure 3.
Assessment of ZFNickase-mediated HDR using a human cell-based chromosomal EGFP reporter assay. (a) A schematic of the U2OS.LacZ-HX735-∂GFP reporter construct integrated in a U2OS cell line. Note that the orientation of the binding site in the reporter is inverted relative to the configuration at the HX735 endogenous locus, for which the Left and Right designations were originally (but arbitrarily) assigned. (b) ZFN and ZFNickase-mediated HDR in a U2OS EGFP reporter line. Cells were co-transfected with the donor plasmid and plasmids encoding HX735 ZFN pairs composed of active Left/active Right (+/+), inactive Left/active Right (−/+), active Left/inactive Right (+/−) and inactive Left/inactive Right (−/−) FokI domains. The graph shows the percentage of EGFP-positive cells 8 days following transfection. Statistically significant differences in HDR-based gene correction relative to donor-only control (cto) are indicated by * (P < 0.05) or ** (P < 0.01).