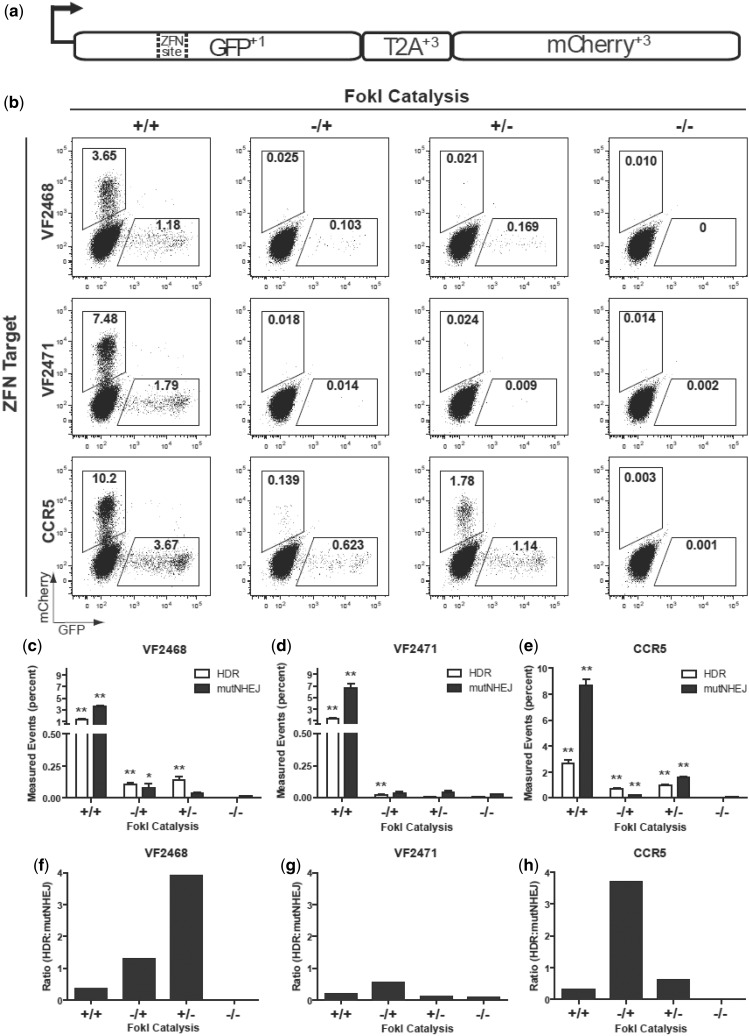
Figure 4.
Assessment of ZFNickase-mediated HDR and NHEJ using a human cell-based TLR assay. (a) Schematic of the ‘TLR’ HDR-mediated correction of the EGFP gene with a co-transfected donor template results in EGFP-positive cells. Mutagenic NHEJ events at the nuclease target site result in mCherry-positive cells. (b) Representative flow cytometry plots showing percentages of EGFP-positive and mCherry-positive cells following transfection of TLR cell lines with plasmid encoding the indicated ZFNs and ZFNickases and the donor template. In the experiments shown, cells have been gated for BFP expression (encoded by the plasmid harboring the donor template) to normalize for transfection efficiencies. (c–e) Bar graphs showing mean percentages of EGFP-positive and mCherry-positive cells for experiments performed with the VF2468, VF2471 and CCR5 ZFNs and ZFNickases. Results were derived from three independent experiments with SEM shown. Statistically significant differences in HDR and mutagenic NHEJ rates relative to donor-only control (−/−) are indicated by * (P < 0.05) or ** (P < 0.01). (f–h) Ratios of percentage of EGFP-positive cells to percentage of mCherry-positive cells for the VF2468, VF2471 and CCR5 ZFNs and ZFNickases using the data from (c–e). Data used to create Figure 4c–h is available in Supplementary Table S3.