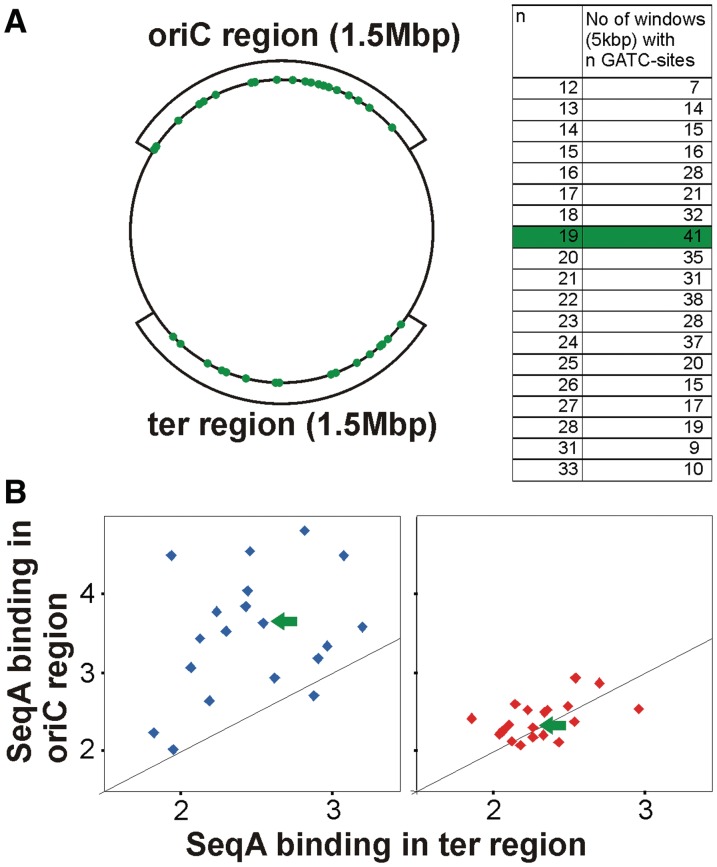
Figure 6.
New replication forks are more frequently bound by SeqA. (A) The chromosome was divided in 5 kbp windows and grouped according to their GATC content and the number of respective windows in two 1.5 Mbp regions at the origin and terminus was calculated as indicated. An example of 41 windows with 19 GATCs is marked in green. (B) The average SeqA ChIP signal was calculated for all windows in the ter or oriC region with the same number of GATC sites. Values were plotted versus each other for data derived from cells grown in LB-glucose (blue) and AB-acetate (red). The black line indicates the theoretical pattern if binding is the same in the oriC and the ter region.