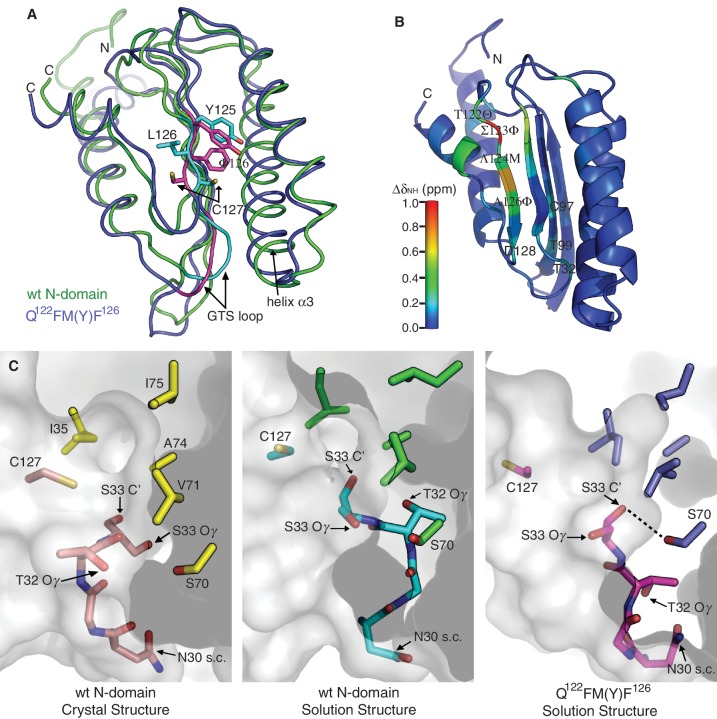
Figure 1.
Structural comparison between wt N-domain and Q122FM(Y)F126. (A) Superposition of the solution structures of wt N-domain (green) and Q122FM(Y)F126 (blue). Regions that are structurally distinct between the two are highlighted in cyan (wt N-domain) and magenta (Q122FM(Y)F126). (B) Differences in amide chemical shift between wt N-domain and Q122FM(Y)F126 are calculated by ΔδNH = [(ΔδH)2 + (0.14*ΔδN)2]1/2, and are mapped onto the structure of wt N-domain according to the colour scale. The four mutated residues are denoted in italics. (C) Conformations of the GTS loop (residues N30–S33) found in the crystal structure of wt N-domain (left panel), the solution structure of wt N-domain (middle panel), and the solution structure of Q122FM(Y)F126 (right panel). Positions of the residues that form the hydrophobic core above the GTS loop are shown by their side-chains only. Hydrogen bonding between the hydroxyl group of S70 and the carbonyl oxygen of S33 in Q122FM(Y)F126 mutant is denoted by a dashed line. The distance between the hydrogen donor and the acceptor is 2.67 ± 0.09 Å.