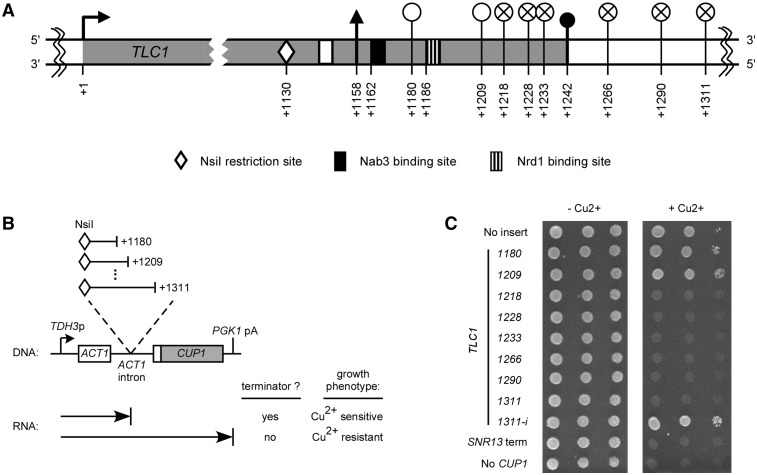
Figure 2.
Sequences required for transcription termination upstream of the Tlc1 polyadenylation sites. (A) Schematic representation of the TLC1 transcription unit with a focus on the 3′-end. The numbers are relative to TLC1 transcription start site (+1). The white circles (empty or crossed) correspond to 3′-end positions of the various TLC1 sequences cloned in the ACT1–CUP1 reporter gene represented in (B). Empty white circles represent constructs containing a sequence that allows transcription read-through and crossed circles represent constructs carrying a sequence that cause transcription termination. See Figure 1 for symbols representing transcription start, polyA− 3′-end, polyA+ 3′-end and Sm protein binding sites. (B) Diagram of the ACT1–CUP1 fusion reporter gene with cloned intronic Tlc1 inserts and the expected phenotypes on copper plates for presence/absence of a terminator element. (C) Tenfold serial dilutions of a culture with the cup1Δ strain 46a containing ACT1–CUP1 constructs with no insert or with inserted sequences from Tlc1 3′-end. The cloned sequences start at the NsiI site at position +1130 and end at position +1180, +1209, +1218, +1228, +1233, +1266, +1290 or +1311, relative to transcription start site (+1). The different cloned TLC1 3′-end sequences are identified by their last nucleotide position. The TLC1–1311–i construct represents the +1130 to +1311 region cloned in the inverted orientation. The SNR13 snoRNA terminator is used as a positive control for transcription termination (SNR13 term). A strain harbouring no ACT1–CUP1 construct (but containing an empty LEU2 plasmid) is used as a control for copper sensitivity (No CUP1). Cultures were spotted on medium lacking leucine and containing 0 or 0.5 mM added copper and incubated for 3 days at 30°C.