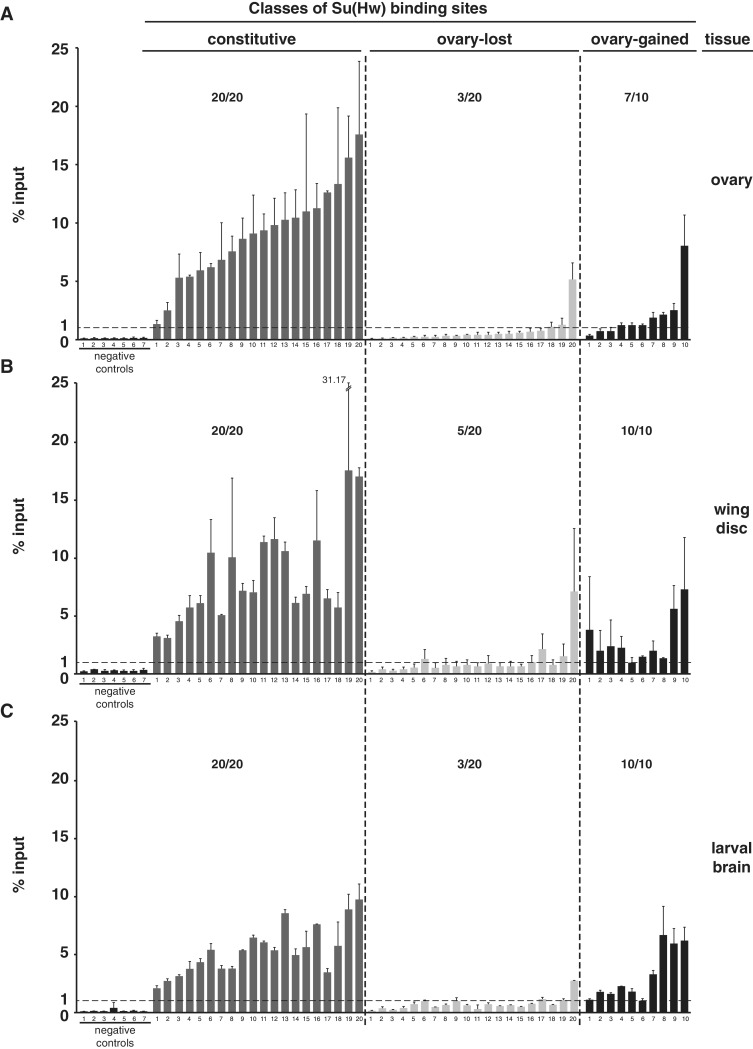
Figure 2.
Su(Hw) binding is largely constitutive. (A–C) Shown is ChIP-qPCR data analyzing Su(Hw) binding to the same sets of SBSs in chromatin obtained from the ovary (A), third instar wing disc (B) and third instar larval brain with eye and antennal disc (C). Negative controls represent sites that lack a Su(Hw)-binding motif and were not identified in the ChIP-Seq dataset. The dashed line indicates 1% input threshold. SBSs tested are divided into constitutive (20 sites), ovary-lost (20 sites) and ovary-gained (10 sites). Averaged values of two biological replicates are shown, error bars indicate standard deviation. For each experiment, the ratio represents the number of SBS above 1% input level over the total number of sites tested.