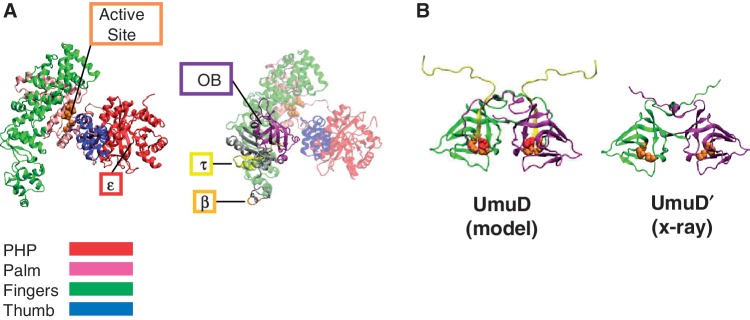
Figure 1.
Models of α and UmuD. (A) The X-ray crystal structure of α 1–917 [PDB code 2hnh (11)] (left) shows the major domains of the polymerase, with the active site and the binding site for ε indicated (23). A homology model of full-length α (24) (right) depicts the C-terminal domain that is not present in the crystal structure containing the β (25) and τ (26) binding sites as well as the OB domain (11,17). (B) The homology model of UmuD (27) and the X-ray crystal structure of UmuD′ (28) showing one monomer in green and the other in purple. The first 24 N-terminal residues missing from the UmuD′ structure (yellow) are cleaved as part of a primitive DNA damage checkpoint by the active site (orange). The single cysteine residues at C24 are shown (red) in full-length UmuD.