Table 2.
Phe-tRNAPhe variant parametersa
| E. coli | E. coli Cy3 | E. coli Cy5 | E. coli Cy5.5 | Yeast | Yeast Cy3 | Yeast Cy5 | |
|---|---|---|---|---|---|---|---|
| Phe-tRNAPhe preparation (%) | |||||||
| Labeling | 80 | 90 | 79 | 92 | 93 | ||
| Charging | 28 | 25 | 15 | 26 | 44 | 37 | 30 |
| Empirical parameters | |||||||
| tLb (2 µM) | 1.00 ± 0.05 | 1.47 ± 0.11 | 1.13 ± 0.22 | 1.13 ± 0.08 | 1.20 ± 0.11 | 1.27 ± 0.11 | 2.33 ± 0.15 |
| tLb (4 µM) | 1.27 ± 0.11 | 1.67 ± 0.04 | 1.07 ± 0.08 | NA | 1.07 ± 0.07 | 1.18 ± 0.11 | 2.27 ± 0.25 |
| S1c (2 µM) | 1.00 ± 0.22 | 0.48 ± 0.17 | 0.42 ± 0.04 | 0.71 ± 0.19 | 0.79 ± 0.04 | 0.42 ± 0.02 | 0.21 ± 0.02 |
| S1c (4 µM) | 2.34 ± 0.06 | 0.60 ± 0.07 | 0.43 ± 0.04 | NA | 1.43 ± 0.05 | 0.50 ± 0.03 | 0.09 ± 0.01 |
| A1d (2 µM) | 1.00 ± 0.09 | 0.87 ± 0.10 | 0.35 ± 0.05 | 0.55 ± 0.17 | 1.02 ± 0.04 | 0.68 ± 0.01 | 0.38 ± 0.02 |
| A1d (4 µM) | 2.89 ± 0.06 | 1.52 ± 0.09 | 0.81 ± 0.03 | NA | 2.41 ± 0.01 | 1.24 ± 0.06 | 0.50 ± 0.02 |
| S2e (2 µM) | 1.00 ± 0.14 | 1.17 ± 0.15 | 0.63 ± 0.07 | 0.37 ± 0.18 | 1.14 ± 0.08 | 0.71 ± 0.09 | 0.68 ± 0.04 |
| S2e (4 µM) | 2.61 ± 0.12 | 1.54 ± 0.12 | 0.78 ± 0.11 | NA ± | 1.74 ± 0.03 | 1.03 ± 0.05 | 0.73 ± 0.09 |
| Relative 14C-Phe incorporationf | |||||||
| 1.00 ± 0.02 | 0.98 ± 0.02 | 0.13 ± 0.01 | NA | 0.66 ± 0.01 | 0.43 ± 0.02 | 0.12 ± 0.01 | |
| Simulation | |||||||
Relative 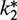
|
1g | 0.1 | 0.025 | 0.2 | 1g | 0.05 | 0.025 |
| Phase 2 steady-state Phe-tRNAPhe concentration (µM) | 0.1 | 0.1 | 0.01 | 0.01 | 0.15 | 0.025 | 0 |
aData at 2 µM added Phe-tRNAPhe are presented in Figure 4B and C. Parallel results obtained at 4 µM Phe-tRNAPhe are summarized in the table. Experiments were performed using 2–5 repeats. Data obtained with 2 µM E. coli tRNA variants was repeated with two different CFPS batches. All values were normalized to 2 µM E. coli Phe-tRNAPhe.
bLag time.
cPhase-1-slope.
dPhase-1-amplitude.
ePhase-2-slope.
fAfter 30-min incubation.
gEqual to k2.
