Figure 1.
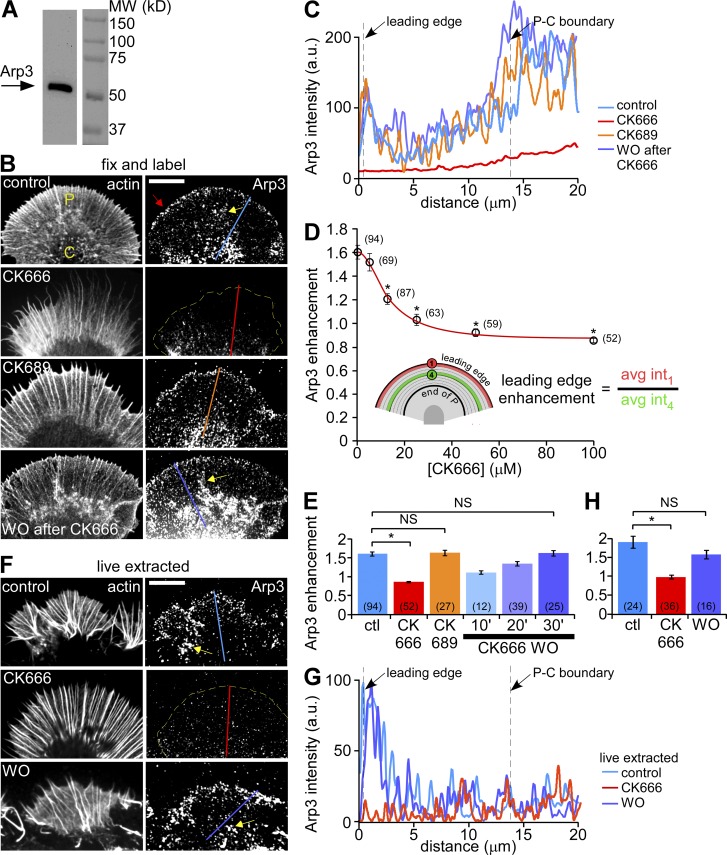
Arp2/3 complex inhibition disrupts its localization in growth cones. (A) Western blot analysis of Aplysia CNS proteins with anti-Arp3 antibody. (B) Fluorescent labeling of Aplysia bag cell neuron growth cones with Arp3 antibody and Alexa 594 phalloidin after normal fixation. Growth cones were treated with vehicle (DMSO, top panel), CK666 (100 µM, second panel), the inactive analogue CK689 (100 µM, third panel) for 20 min, or treated with CK666 (100 µM) for 20 min followed by 30 min recovery in control medium (bottom panel). Red arrow: Arp2/3 complex enrichment. (C) Arp3 distribution profile sampled from the designated lines in B. (D) CK666 dose-dependent reduction of Arp2/3 complex enrichment at the leading edge. Red line: best fit curve using the 4-parameter nonlinear regression model (see Materials and methods), R2 = 0.9929. *, P < 0.01 with two-tailed unpaired t test versus the first data point (control). (E) Quantification of Arp2/3 complex enrichment at the leading edge for each condition in B. For images of 10- and 20-min CK666 washout, see Fig. S2 C. *, P < 0.01 with two-tailed unpaired t test. NS, not significant. (F) Fluorescent labeling of growth cones with Arp3 antibody and TRITC-phalloidin after live cell extraction. Growth cones were treated with vehicle (DMSO, top), CK666 (100 µM, middle) for 20 min, or treated with CK666 (100 µM, 20 min) followed by recovery in control medium for 30 min (bottom). (G) Arp3 distribution profiles sampled from the designated lines in F. (H) Quantification of Arp2/3 complex enrichment at the leading edge for each condition in F. *, P < 0.01 with two-tailed unpaired t test. NS, not significant. Yellow dotted lines demarcate the leading edge. Yellow arrows, intrapodia. Numbers in parentheses indicate growth cones measured. Bars, 10 µm.