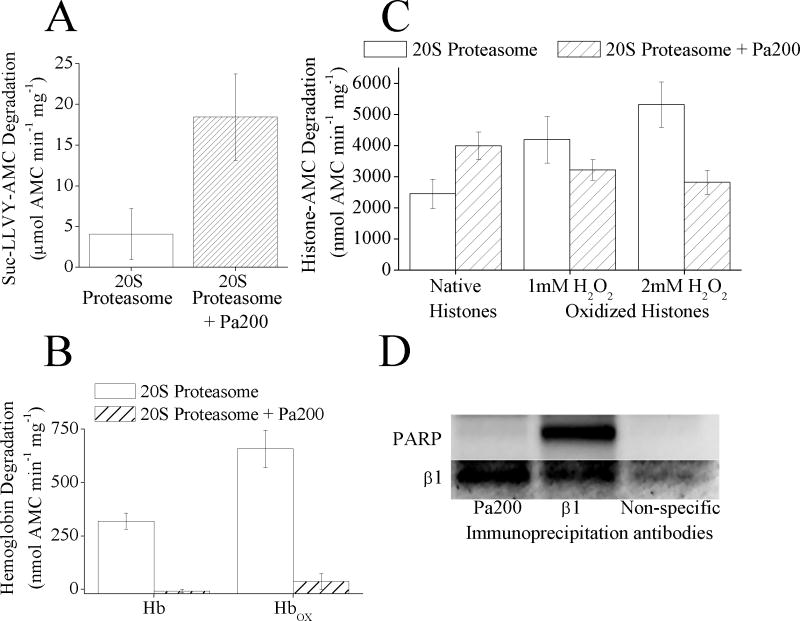
Fig. 4.
(A) Pa200 increases the capacity of 20S proteasome to degrade the peptide substrate Suc-LLVY-AMC. 20S proteasome was incubated with a 4 fold molar excess of Pa200 for 30 minutes. Suc-LLVY-AMC was then added to samples and proteolytic capacity measured over the next 4 h. Values are means ± SE where n = 3. (B) Pa200 reduces the capacity of 20S proteasome to degrade both oxidized and naive hemoglobin substrates. 20S Proteasome ± Pa200 was prepared as in panel A. Hb-AMC or HbOX-AMC was then added to samples and proteolytic capacity measured over the next 4 h. Values are means ± SE, where n = 8. (C) Pa200 enhances the capacity of proteasome to degrade normal histones but reduces proteasome’s capacity to degrade oxidized histones. 20S Proteasome ± Pa200 was prepared as in panel A and AMC labeled normal histones, or oxidized AMC labeled histones were used as substrates. Oxidized histones were prepared by treatment with either 1mM or 2mM H2O2 for 1 h. Proteolysis values are means ± SE, where n = 6. (D) Immunoprecipitation with either anti-Pa200 or anti-β1 both result in co- immunoprecipitation of 20S proteasome subunits, however only anti-β1 antibodies co-precipitate PARP. MEF cells were grown to 10% confluence, then immunoprecipitation with either anti-β1 antibody, anti-Pa200 antibody, or anti-porin antibody (as a non-specific binding control) was performed. Samples were run on Western blots and screened for co-immunoprecipitation of anti-β1 or anti-PARP.