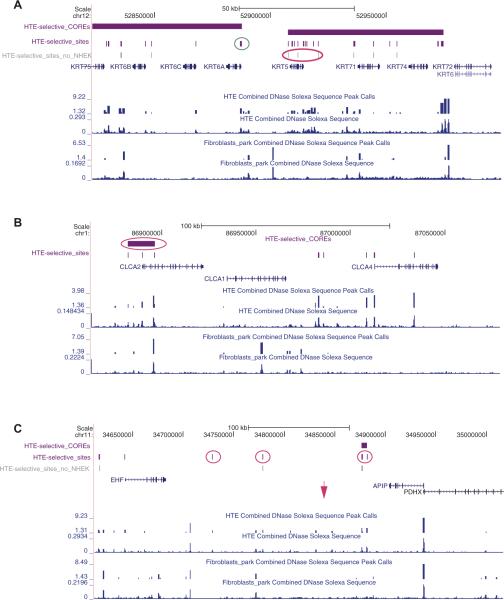
Figure 4.
COREs of open chromatin are associated with critical genes in human tracheal epithelial cell function. Each panel (A–C) shows a region of the human genome on the University of California Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu) with combined DNase Solexa (Illumina) sequence data and peak calls for two biological replicates of human tracheal epithelial (HTE) cells and skin fibroblasts (Fibroblasts_park). UCSC genes within the region are also shown. Above these tracks are HTE-selective COREs (horizontal purple bars), HTE-selective DNase I hypersensitive sites (DHS) (vertical purple bars) and (in A, C) HTE-selective sites that do not overlap with sites in normal human epidermal keratinocytes (NHEK) (vertical grey bars, HTE-selective_sites_no_NHEK). Pink and teal circles show HTE-selective COREs or DHS of particular interest (see text). (A) Part of the type II cytokeratin cluster on chromosome 12q12-q13 showing the regions of open chromatin around the keratin 5 (KRT5) and 6A (KRT6A) genes, which are both highly expressed in HTE cells. (B) The calcium-sensitive chloride channel accessory protein (CLCA) cluster on chromosome 1p22. CLCA2 is highly expressed in HTE cells, CLCA1 is not. (C) The EHF–APIP (Ets homologous factor, epithelial specific–APAF1-interacting protein, anti-apoptotic) interval which shows strong association with lung disease severity in a cystic fibrosis modifier genome-wide association study. The pink arrow shows the location of the single nucleotide polymorphism with the highest association. PDHX, pyruvate dehydrogenase complex, component X.