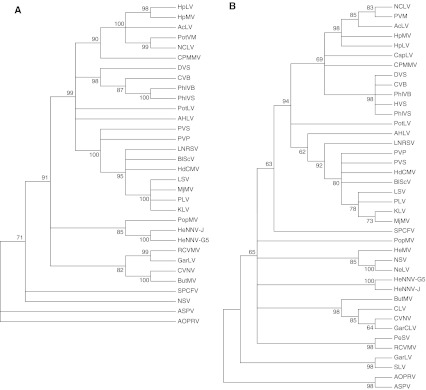
Fig. 1.
Cladogram of the predicted amino acid sequences from the replicase (A), and coat protein (B) of members of the genus Carlavirus. The phylogenetic tree was generated using the maximum-likelihood method in the MEGA5 analysis package. Branch significance was evaluated by constructing 1,000 trees in bootstrap analysis, and the bootstrap values (>60 %) are shown. The sequences used were derived from the genomic sequences of Aconitum latent virus (AcLV, gi: 14251190), American hop latent virus (AHLV, gi: JQ245696), blueberry scorch virus (BlScV, gi: 19919919), butterbur mosaic virus (ButMV, gi: 269954638), chrysanthemum virus B (CVB, gi: 154814487), coleus vein necrosis virus (CVNV, gi: 156616260), cowpea mild mottle virus (CPMMV, gi: 313122763), daphne virus S (DVS, gi: 94971809), garlic latent virus (GarLV, gi: 20143432), Helleborus net necrosis virus (HeNNV-G5, gi: 222509601 and HeNNV-J, gi: 349584640), hop latent virus (HpLV, gi: 10314012), hop mosaic virus (HpMV, gi: 171473627), hydrangea chlorotic mottle virus (HdCMV, gi: 242346753), Kalanchoe latent virus (KLV, gi: 254667440), Ligustrum necrotic ringspot virus (LNRSV, gi: 166851938), lily symptomless virus (LSV, gi: 37674199), Mirabilis jalapa mottle virus (MjMV, gi: 352749693), narcissus common latent virus (NCLV, gi: 110835685), narcissus symptomless virus (NSV, gi: 116734779), Passiflora latent virus (PLV, gi: 113195533), phlox virus B (PhlVB, gi: 160700642), phlox virus S (PhlVS, gi: 145651764), poplar mosaic virus (PopMV, gi:4 1057345), potato latent virus (PLV, gi: 211998869), potato virus M (PVM, gi: 72257063), potato virus P (PVP, gi: 156603882), potato virus S (PVS, gi: 71849435), red clover vein mosaic virus (RCVMV, gi: 224923880), and sweet potato chlorotic fleck virus (SPCFV, gi: 56692800). In addition, the following carlaviruses were included in the analysis of coat protein sequences: caper latent virus (CapLV, gi: 333034471), cole latent virus (CLV, gi: 33517421), garlic common latent virus (GarCLV, gi: 3077682), Helenium virus S (HVS, gi: 221446), Helleborus mosaic virus (HeMV, gi: 222142735), Nerine latent virus (NeLV, gi: 298504072), pea streak virus (PeSV, gi: 13811416), shallot latent virus (SLV, gi: 3077693). The corresponding sequences of apple stem pitting virus (ASPV, gi: 19744938) and African oil palm ringspot virus (AOPRV, gi: 226202432) were used as out-groups