Figure 3.
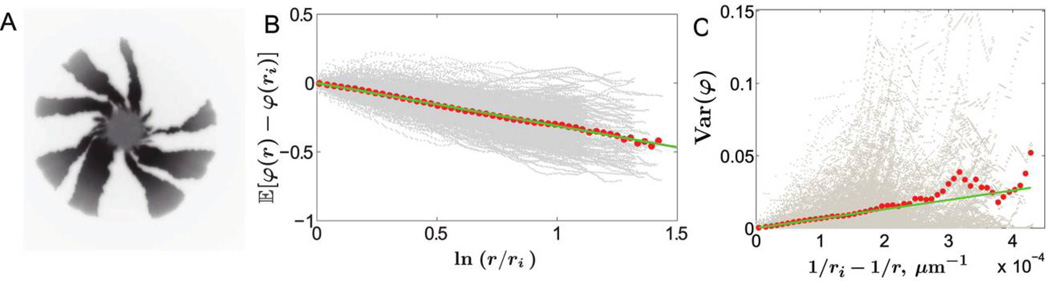
Chirality and boundary wandering in Escherichia coli colonies. A, Genetic demixing in a chiral E. coli colony. The image, taken from the bottom of the petri dish, shows the fluorescent signal from only one of the two segregating alleles. Bacterial colonies of Pseudomonas aeruginosa at 37°C and 21°C did not exhibit chiral growth. B, Twisting of sector boundaries in E. coli colonies. The gray dots represent individual measurements along the sector boundaries at pixel resolution (one pixel is 50 µm) from 30 colonies (390 sector boundaries). Each red dot is the mean position of the gray dots in one of the 50 equidistant bins along the X-axis. The green line is the least square fit to the red dots. According to equation (9), the slope of the green line equals v⊥/v‖, which yields v⊥/v‖ = 0.32. As shown in A and B, all sector boundaries twist on average in the same direction for chiral growth. In contrast, sector boundaries between nonneutral strains bend in both clockwise and counterclockwise directions because the boundaries around the fitter strains bend outward (Hallatschek and Nelson 2010). To exclude possible small fitness differences, we also separately analyzed boundaries that would bend in opposite directions if the strains were nonneutral. Nonneutrality would result in different values of v⊥/v‖ for these two sets of boundaries. Upon applying equivalent tests to those used in figure 5, we confirmed that v⊥/v‖ are the same, which supports the previous finding that these strains are neutral (Hallatschek et al. 2007). C, Random walks of sector boundaries. The same data as in B are used to plot the variance of φ(r); gray dots are the individual data points, and red dots are the averages. The green line is the least square fit to the first 25 red dots; the last 25 dots are not used because of large fluctuations due to a smaller sample size at large r. According to equation (10), the slope of the green line equals 2Ds/v‖. The data sets for P. aeruginosa at 37°C and 21°C show similar behavior of the variance of φ(r), but the data set for E. coli fluctuates less partly because of the larger sample size and larger spatial diffusion constant compared with those of the other two experiments.