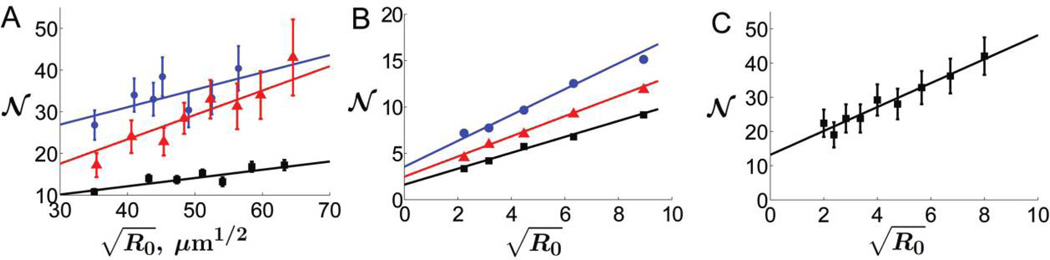
Figure 6.
Dependence of diversity at the expansion front on the initial size of the population in experiments (A), on-lattice simulations (B), and off-lattice simulations (C). The average number of sectors at the end of a range expansion 𝒩 is plotted against the square root of the initial radius of the colony R0. These coordinates are chosen so that the theoretically predicted dependence (see eq. [7]) is a straight line (solid lines). The error bars represent 95% confidence intervals. Parameters Ds/v‖ and Dg/v‖ can be estimated from the slope of the fit and its intercept with the Y-axis, respectively. A, Escherichia coli (black squares), Pseudomonas aeruginosa at 37°C (red triangles), and P. aeruginosa at 21°C (blue dots). B, Three different island-carrying capacities: N = 3 (black squares), N = 30 (red triangles), and N = 300 (blue dots). Each data point is the average of 40 simulations. Notice that the populations with higher N have more sectors in agreement with equation (3) because populations with higher densities have lower Dg. In this plot, the error bars are represented by the size of the markers used. C, Because of the discrete nature of these simulations, we use the square root of the number of cells at inoculation as a proxy for the initial radius.