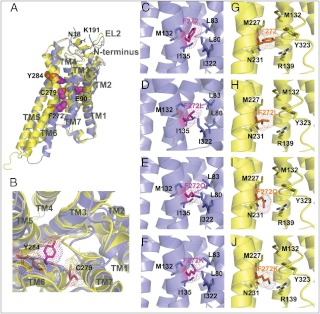
Fig. 5.
Location of mutations in the hGnRHR. Helices are shown by cartoons colored blue (inactive conformation) and yellow (active conformation). Substituted residues are shown by sticks, highlighted by dots, and colored purple (inactive conformation) and orange (active conformation). A, Positions of residues mutated in this work (E90K, F272Q, K, C279A, S, Y284F, and ΔK191) in inactive and active conformations of hGnRHR. Side chains of substituted residues (E90, F272, C279, and Y284) are shown by balls colored purple (for the inactive state) and orange (for the active state). K191 from the EL2 and proximal N18 from the N terminus are shown by sticks colored blue. B, Suggested changes in the ligand binding pocket on receptor activation. Positions of mutated residues (C279 and Y284) in the binding pocket of active (yellow) and inactive (blue) receptor conformations. Helix shift and rotamer change of Y284 and C279 during the receptor activation bring Y284 to the nonpolar lipid environment and C279 to the polar environment of the ligand binding pocket. Note the contraction of the ligand binding pocket in the active state caused by the inward movement of TM5, TM3, and TM7. C–J, Interactions between substituted residues in position 272 from the moving TM6 and adjacent residues from TM3, TM5, and TM7 in the model of the inactive and activated hGnRHR. Environment of F272 of hGnRHR (C and G) and its substitutions, F272L (D and H), F272Q (E and I), and F272K (F and J), in inactive (C–F, blue) or active (G–J, yellow) receptor states.