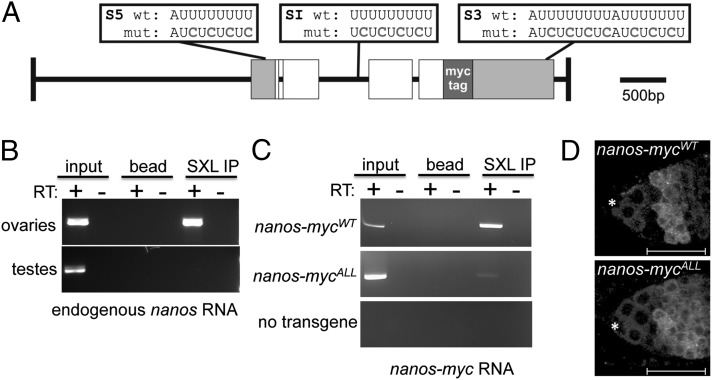
Fig. 2.
The nanos gene is a Sxl target gene. (A) Schematic representation of the 5.8-kb genomic myc-tagged nanos constructs. Gray boxes indicate the 5′ and 3′ UTRs, white boxes represent the ORFs, and green box indicates the 6× MYC-tag inserted in frame. The location of the wild-type and mutated Sxl binding sites are shown above the diagram. (B) Sxl associates with nanos RNA. RNA IP assays were carried out in whole-cell extracts from ovaries or testis. The presence of nanos RNA in the IP pellet was detected by RT-PCR. (C) Mutagenesis of the S5, S1 and S3 binding sites impairs Sxl binding to nanos RNA. RNA IP assays were carried out in whole-cell extracts from ovaries carrying either the WT nanos-mycWT construct, the mutant nanos-mycALL construct, or no transgene. The presence of nanos-myc RNA in the IP pellet was detected by RT-PCR. (C) Confocal images of ovaries carrying either a WT or mutant myc-tagged nanos construct stained for Myc. The staining pattern of the nanos-mycWT parent construct resembles WT, whereas the mutant nanos-mycALL construct in which all of the S5, SI, and S3 binding sites are mutated is not down-regulated in the early differentiating germline cells. The location of the somatic cap cells is marked with an asterisk, and the regions are marked as in Fig. 1. Scale bars = 25 μm.