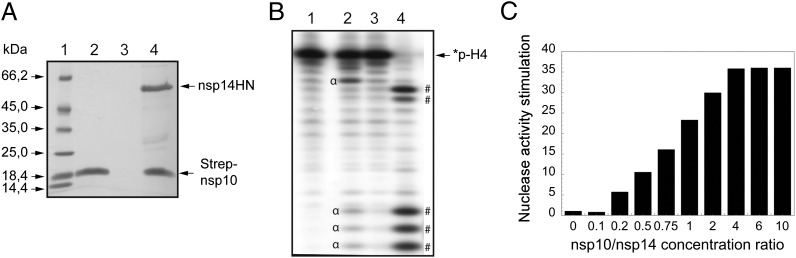
Fig. 1.
Nsp10 interacts with nsp14, and nsp10/nsp14 shows enhanced ExoN activity. (A) SARS-CoV nsp14HN and Strep-nsp10 proteins coexpressed or expressed alone were incubated with Strep-Tactin Sepharose. Strep-Tactin–bound proteins were eluted with d-desthiobiotin and analyzed by SDS/PAGE and Coomassie blue staining. Lane 1 corresponds to the molecular size markers; lane 2 to Strep-nsp10 expressed alone; lane 3 to nsp14HN expressed alone; and lane 4 to Strep-nsp10 coexpressed with nsp14HN. (B) Autoradiogram of RNA cleavage products. Synthetic *p-H4 RNA was radiolabeled at its 5′-end using PNK in the presence of [γ32P]ATP (the asterisk indicates the 32P-labeling position). *p-H4 RNA was incubated at 37 °C in Tris⋅HCl buffer 40 mM (pH 8), DTT 5 mM with no protein (lane 1), 0.7 μM of nsp14 (lane 2), nsp10 (lane 3), or both proteins (lane 4) during a 90-min period. The reaction products were then separated on a 20% (wt/vol) denaturing Urea-PAGE and revealed using photostimulated plates and a FujiImager (Fuji). (C) *p-H4 RNA was hydrolyzed with fixed nsp14 concentration (50 nM) in the presence of increasing concentration of nsp10 ranging from 0 to 1,600 nM (nsp10/nsp14 is indicated below the bar graph). ExoN activity was quantified using denaturing Urea-PAGE followed by measuring the hydrolysis of *p-H4 corresponding band using a FujiImager and Image Gauge software analysis.