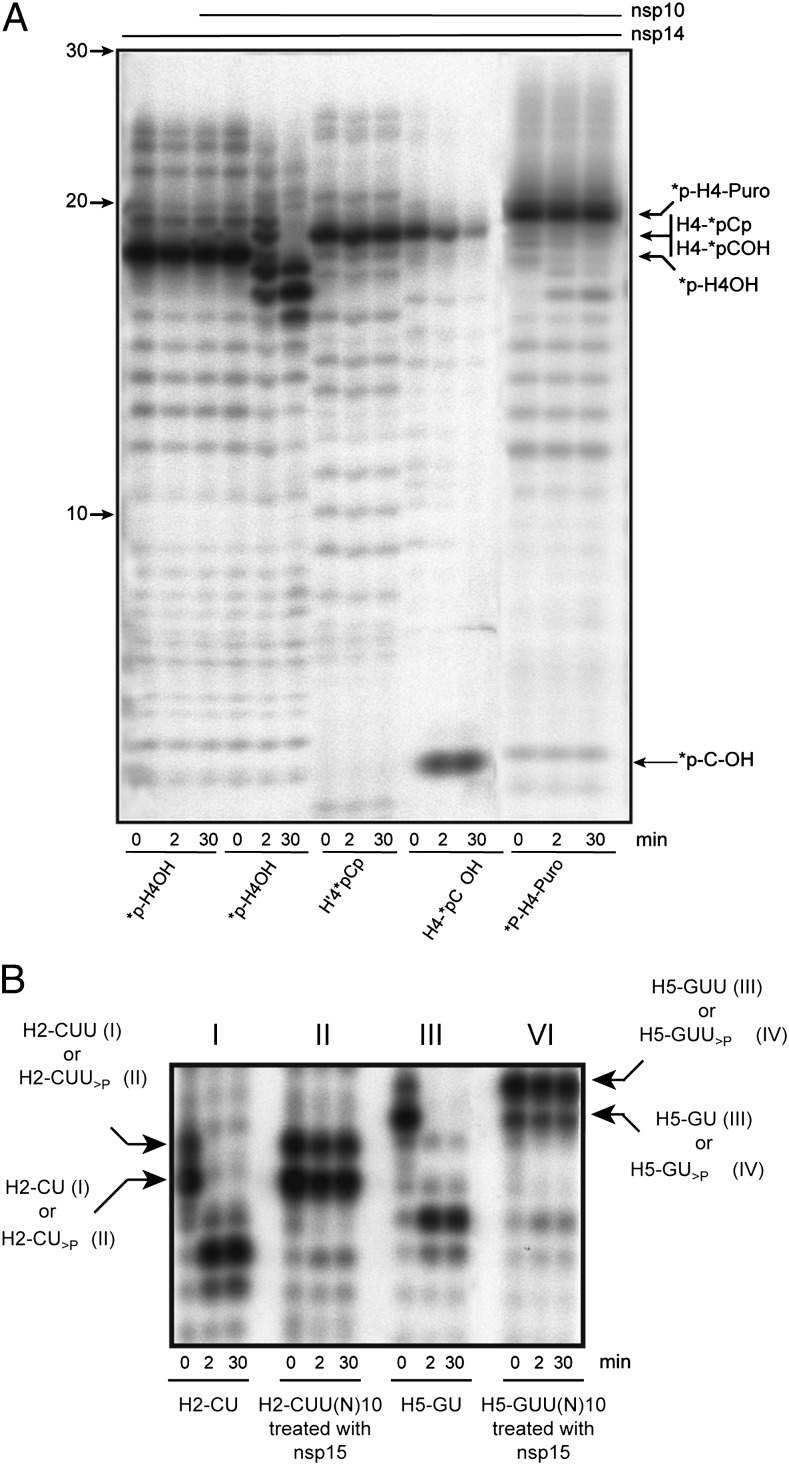
Fig. 5.
Nsp10/nsp14 requires a free 3′-hydroxyl end. (A) Autoradiogram of an ExoN assay performed with RNA carrying 3′-phosphate, 3′-puromycin, or 3′-hydroxyl end. The 3′ modified RNAs (*p-H4 as control, *p-H4-puromycin, H4-*pCp, and H4-*pCOH) were incubated with nsp14 or nsp10/nsp14 for 0, 2, and 30 min (asterisk indicates the 32P-labeling position). Digestion products were separated on 20% (wt/vol) Urea-PAGE and revealed by autoradiography. (B) Autoradiogram of nsp15-generated 2′-3′-cyclic phosphate RNAs cleavage assay. RNA substrates H2-CUU(N)10 and H5-GUU(N)10 were synthesized using T7 RNA polymerase and subsequently dephosphorylated and radiolabeled. These RNAs were then incubated with nsp15 to generate *p-H2-CU>P and *p-H2-CUU>P and, to a lesser extent, *p-H5-GUU>P and *p-H5-GU>P. Control RNAs *p-H2-CUOH and *p-H5-GUOH were synthesized using T7 RNA polymerase and led to the production of the side products *p-H2-CUUOH and *p-H5-GUUOH, respectively. After purification, these substrates were incubated with nsp10/nsp14 for 0, 2, and 30 min. Digestion products were separated on 20% (wt/vol) Urea-PAGE and revealed by autoradiography.