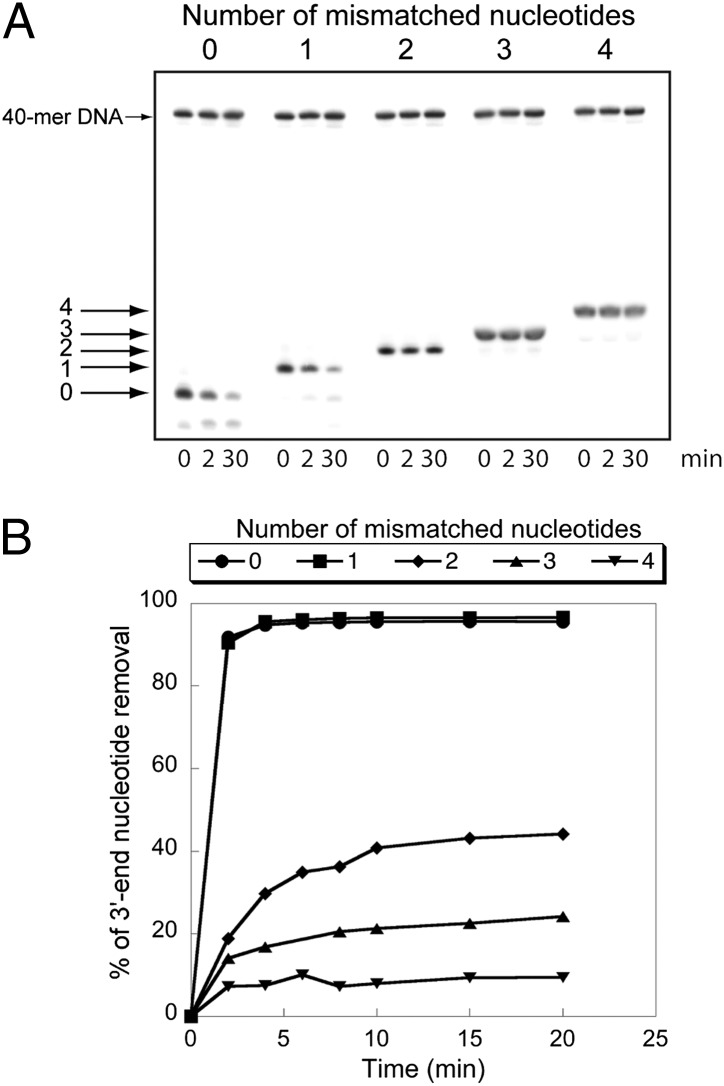
Fig. 6.
Comparison of nsp10/nsp14 ExoN activity on paired and mismatched 3′-end nucleotide base pairs. (A) A 40-nt RNA (LS1) was annealed with 5′-radiolabeled oligoribonucleotides carrying zero, one, two, three, or four noncomplementary bases at its 3′-end (respectively, LS2, LS3, LS4, LS5, or LS6 (Table S1 and Fig. S5). To avoid nsp10/nsp14-mediated LS1 degradation, this RNA carries a biotin group at its 3′-end. Duplex RNAs were then incubated (0, 2, and 30 min) with purified nsp10/nsp14 (200 nM/50 nM). Reaction products were separated on a 20% (wt/vol) denaturing Urea-PAGE and revealed by autoradiography. A radiolabeled 40-mer DNA was introduced in the reaction mixture as a quantification reference. (B) Kinetics of mismatch excision. The assay was performed as in A, using 100 nM of nsp14 and 400 nM of nsp10. RNA cleavage was quantified at 0, 2, 4, 6, 8, 10, 15, and 20 min. Data are presented as percent of 3′-nucleotide removal.