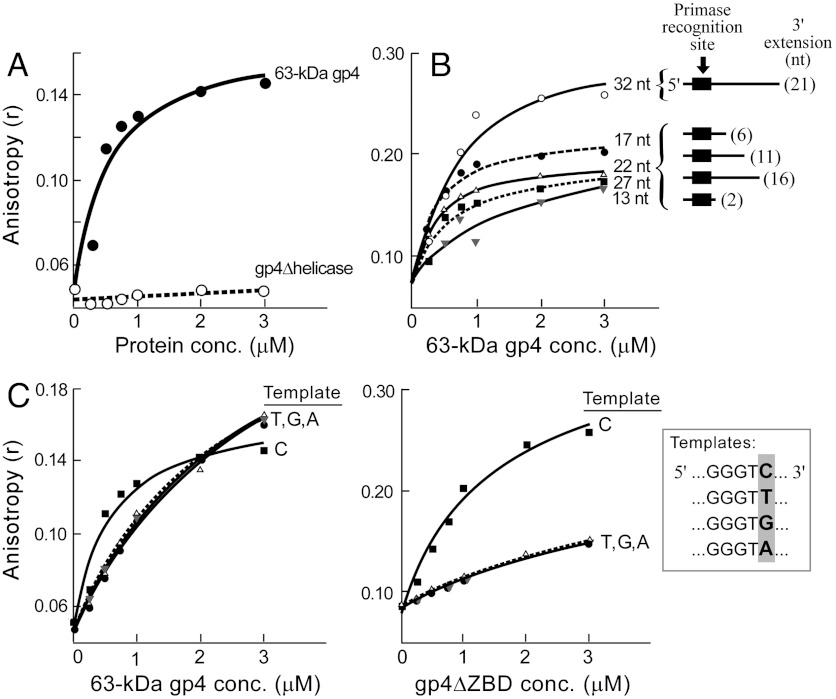
Fig. 5.
The role of the helicase domain of gp4, and the “cryptic” C in the stabilization of the priming complex. (A) The helicase domain of gp4 stabilizes the priming complex. Reaction mixtures contained the indicated concentrations of 63 kDa gp4 (filled circles) or gp4∆helicase (open circles), 2 μM gp5/trx, 0.1 μM template (5′-CAG TAG CGG GTC TAT TTC TCA GCG TCG-6-FAM-3′), and 0.5 μM primer (5′-rArCrCrCdGddC-3′). After 30 min incubation at 25 °C anisotropy was measured, and the anisotropy values are plotted as a function of protein concentrations. (B) Priming complex binding to different templates. Five templates (see Inset) with varying 3′extensions: 21 nt (open circles), 16 nt (filled squares), 11 nt (open triangles), 6 nt (filled circles), and 2 nt (gray triangles) were incubated in a reaction mixture containing 0.5 μM primer (shown above), 2 μM gp5/trx, and increasing concentrations of 63 kDa gp4. Anisotropy was measured after 30 min incubation. (C) Stability of the priming complex using DNA templates (see Inset) in which the cryptic cytosine (template C) was substituted with thymine, guanine or adenine (templates: T, G, or A, respectively). Each template (0.1 μM) was incubated in a reaction mixture containing 0.5 μM primer, 2 μM gp5/trx, and increasing concentrations of 63 kDa gp4 or gp4∆ZBD. Anisotropy readings are shown as a function of increasing concentrations of 63 kDa gp4 or gp4∆ZBD.