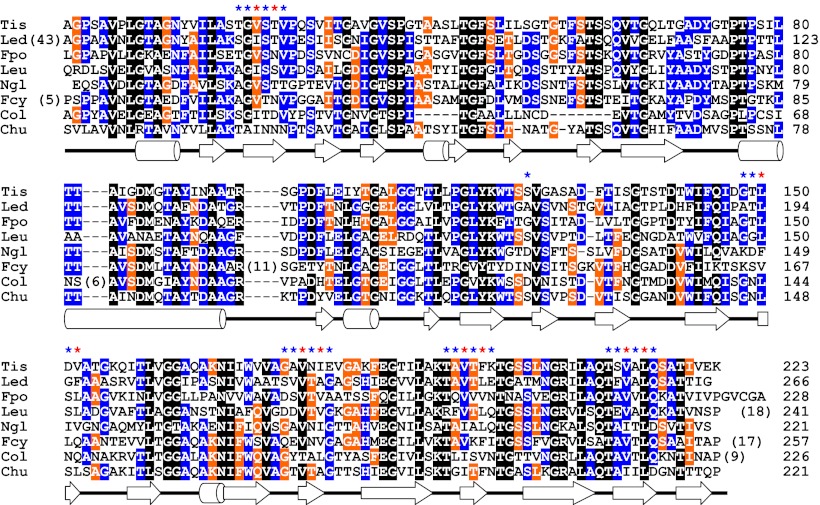
Fig. 5.
Amino acid sequence alignment of TisAFP6 homologs identified in various microorganisms: Tis, Typhula ishikariensis; Led, Lentinula edodes; Fpo, Flammulina populicola; Leu, Leucosporidium sp AY30; Ngl, Navicula glaciei; Fcy, Fragilariopsis cylindrus; Col, Colwellia sp. SLW05; and Chu, Cytophaga hutchinsonii. The positions of α-helices and β-strands in the TisAFP6 structure are indicated below the alignment by cylinders and arrows, respectively. Secondary structure assignments were performed with DSSP (40). Residues conserved between seven or eight species are indicated by a black background, those between five or six species by a blue background, and between four species by an orange background. The asterisks above the alignment show the residues on the putative IBS of TisAFP6. Blue asterisks signify outward pointing residues and orange asterisks signify outward pointing residues. On the Right, numbers indicate the residue number of each polypeptide from the start of the mature protein. Numbers in parentheses indicate the length insertions. Elsewhere, dashes indicate residues missing from one or more homologs.