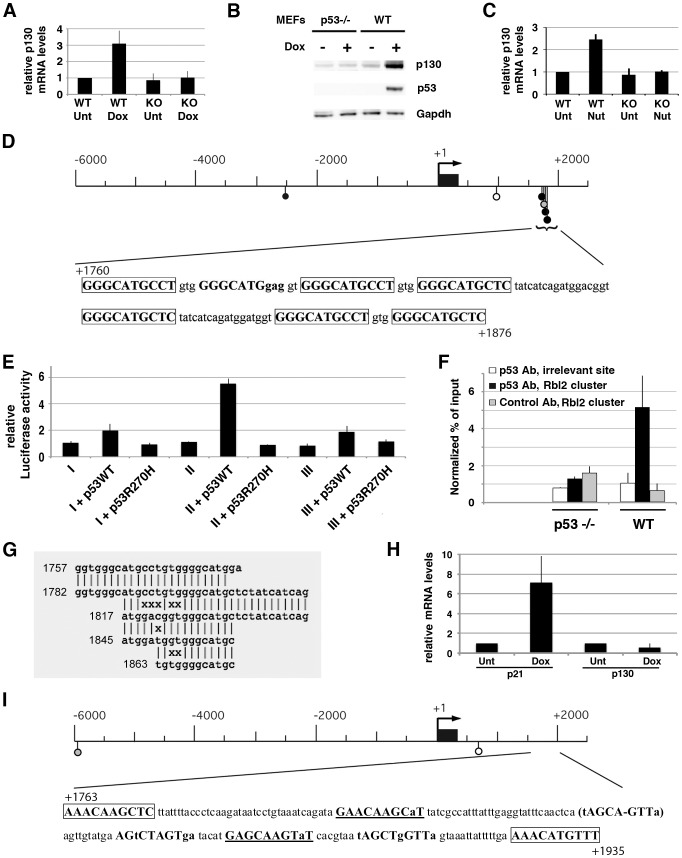
Figure 1. Murine Rbl2/p130 is a p53 target gene.
(A) WT and p53−/− MEFs were left untreated (Unt) or treated with doxorubicin (Dox) before RNA extraction and real-time PCR quantification, in 8 independent experiments. Data were normalized to control mRNA levels, then a value of 1 was assigned to mRNA amounts in unstressed WT cells. (B) MEFs were treated as in (A), then protein extracts were immunoblotted with antibodies to p130, p53 and Gapdh. (C) MEFs were left untreated (Unt) or treated with Nutlin (Nut) before RNA quantification, in 4 independent experiments. (D) Putative p53 REs, identified using Consite and a positional frequency matrix (Methods), were plotted along the Rbl2/p130 locus as lollipops, with greytones according to scores (Table S1). Numbers are relative to the transcription start site (TSS). Black box: exon 1. Below, the cluster sequence is shown, with p53 putative binding half-sites in bold (putative binding sites have 0–3 mismatches with the consensus; matches in capital letters and mismatches in lowercase; perfect half-sites are boxed). (E) The 6 kb upstream the TSS were cloned before a luciferase reporter gene, and the plasmid was transfected in p53−/− MEFs alone (I), or with an expression vector for WT p53 (I+p53WT) or mutant p53 (I+p53R270H) to measure luciferase. Likewise, luciferase was measured with plasmids containing 2.5 kb of sequences downstream the TSS with (II) or without (III) the clustered p53 REs. Results, from 3 independent experiments, were normalized to control renilla luciferase, then a value of 1 was assigned to luciferase in cells transfected with reporter plasmids alone. (F) ChIP assay was performed in doxorubicin-treated MEFs, with an antibody against p53, or rabbit IgG as a control. Immunoprecipitates were quantified using real-time PCR and normalized to input DNA on an irrelevant region, in 3 independent experiments. (G) Integrated results of the cluster sequence analysis with mreps. Numbers are relative to the TSS. (H) Human lung fibroblasts were treated and mRNAs were quantified as in (A), in 3 independent experiments. Similar results were obtained with foreskin fibroblasts. (I) Putative p53 response elements were searched for and plotted along the human Rbl2/p130 locus as in (D). The region homologous to the murine clustered p53 REs is below, with putative half-sites as before (those with a single mismatch are underlined). The parentheses indicate a nonamer that might be a putative half-site with a deletion within the core CWWG motif (the minus sign indicates the deletion), a situation observed in about 5% of p53 binding half-sites [41].