Abstract
In this essay I describe my personal journey from reductionist to systems cell biology and describe how this in turn led to a 3-year sea voyage to explore complex ocean communities. In describing this journey, I hope to convey some important principles that I gleaned along the way. I realized that cellular functions emerge from multiple molecular interactions and that new approaches borrowed from statistical physics are required to understand the emergence of such complex systems. Then I wondered how such interaction networks developed during evolution. Because life first evolved in the oceans, it became a natural thing to start looking at the small organisms that compose the plankton in the world's oceans, of which 98% are … individual cells—hence the Tara Oceans voyage, which finished on 31 March 2012 in Lorient, France, after a 60,000-mile around-the-world journey that collected more than 30,000 samples from 153 sampling stations.
FROM REDUCTIONIST TO SYSTEMIC CELL BIOLOGY
Over the past 20 years, cell biology has moved from sheer morphological and molecular description to the analysis of causal relationships between components, using genetics, molecular biology, and imaging. Recent technological and conceptual advances have begun to move the field toward the understanding of the dynamic organization and complex functions of cells (Hartwell et al., 1999; Karsenti, 2008). This involves computer modeling and analytical mathematic analysis using dynamical parameters gathered using biochemistry and live imaging. A nice article published in this journal actually discussed when modeling can be applied to a cell biology problem (Fletcher, 2011).
I cite just two examples. First, genetics and biochemistry have allowed the unraveling of molecular mechanisms that drive the cell cycle. This has led to the discovery of how positive and negative feedback loops, switches, and time delays (Murray, 1989; Félix et al., 1990; Clarke et al., 1993; Hoffmann et al., 1993; Nurse, 1994) build the cell cycle oscillator. Quantitative models of the cycle built using realistic enzymatic parameters have shown how cycles could indeed emerge from such mechanisms (Chen et al., 2004; Ferrell et al., 2011; Krasinska et al., 2011).
Another example is the mitotic spindle. Again, genetics and biochemistry identified many components of the spindle (Manning and Compton, 2008; Tanaka and Desai, 2008; Walczak and Heald, 2008; Gatlin and Bloom, 2010; Wadsworth et al., 2010; Wordeman, 2010). The importance of microtubule dynamic instability, microtubule-associated proteins, and motors in the organization of microtubules into bipolar spindles was discovered. This led to intuitive models of spindle assembly, such as the search-and-capture and the microtubule-motor self-organization models (Kirschner and Mitchison, 1986; Heald et al., 1996). These models have been tested and quantitatively fleshed out by mathematics and computer simulations (Holy and Leibler, 1994; Surrey et al., 2001; Nedelec et al., 2003; Wollman et al., 2005; Athale et al., 2008), leading to new principles, such as the importance of gradients generated by reaction-diffusion mechanisms (Caudron et al., 2005). A quantitative model integrating all these mechanisms and principles has recently been proposed, indicating how a steady-state dynamic spindle could indeed emerge from the collective effects of local regulation of microtubule dynamics and motor activities (Loughlin et al., 2010).
These examples show how “network patterns”—the way enzymes and substrates interact to generate reaction cascades (positive and negative feedback loops, feedforward loops, reaction diffusion processes based on localized and diffusible enzymes, etc.) govern temporal and spatial order in the cell. Different multiple and collective molecular interaction patterns have various reversibility or irreversibility properties, as well as various timing and spatial properties, that underpin the diversity of cell dynamics, organization, and function. Somehow evolution played around with genetic networks a bit like a Meccano game, and only functional combinations/patterns survived, leading to the present cells (Parter et al., 2007; Kashtan et al., 2009).
This brings us to what is, in my opinion, the most revolutionary part of these new developments in cell biology. Because shape, function, and temporal properties of cellular systems “emerge” from multiple interactions between “agents” (molecules or groups of molecules in this case), we are no longer dealing with a simple causality problem. We are instead facing a “system properties” issue. This calls for an additional and very different approach from studies focusing on single-molecule functions/properties. Mathematical models and numerical simulations have been applied in the cell cycle and mitotic fields and are tools that can establish general “emergence principles,” taking into consideration all previously identified causal links. This, in turn, can explain why and how a functional living unit emerges out of its components. The type of prediction one gets from such “self-organization models” is not the same as that provided by differential equations. Differential equations are deterministic from the outset. Emergent models, because they include a large element of stochasticity, tell us that by putting a certain number of interacting agents together under well-defined conditions, the “system” will evolve toward a certain “dynamical state” (e.g., a spindle). However, it is impossible to predict this before having tested the model with simulations and identified the combinatorial landscape that gives rise to the structure in which you are interested! Cells are not “machines” in the engineering sense of the term: They are self-organized dissipative structures. They have not been “designed”; they just “emerge.” This is why it is necessary to screen for the combination(s) of parameter values that lead to the emergence of order. By doing this, we characterize a “system” and acquire a holistic understanding of cells or subcellular functional parts. To sum up, the reductionist approach addresses quantitative causal chains of events that must be integrated into those more holistic models in order to grasp the full essence of living matter.
FROM SYSTEMIC CELL BIOLOGY TO EVOLUTION AND ECOLOGY
The studies just described show that beyond the simple causal effect that a mutation can have on the function of a structural protein or an enzyme, it is also the collective behavior of an ensemble of gene products that determines cell activities and structures. This is what systems biology calls “networks.” A network is built of nodes and links (e.g., molecules and their activities in relation to each other). Mutations inside a network can affect the overall behavior of the network in various ways. They can either kill the network output altogether if a mutation kills an essential enzyme or just change its configuration and produce a dramatic or minor effect on the output. Hence there is no simple correlation between the effect of mutations and evolution. Indeed, a lot of mutations can be neutral in an unchanging environment (the network may fluctuate inside a parameter space without obvious effect on the output) while having a dramatic effect in a varying environment. Such ideas have led to the formulation of general organization principles of regulatory networks (Milo et al., 2002; Di Ventura and Sourjik, 2011), providing grounds for a new approach to evolution (Kashtan et al., 2009) that is deeply rooted in cell biology.
Theoretical studies on the topology of possible functional regulatory networks, as done by Uri Alon (Milo et al., 2002, 2004; Ronen et al., 2002; Itzkovitz et al., 2003; Rosenfeld and Alon, 2003), have led to a list of intracellular network patterns and properties that may exist in living systems and shown how networks can switch from one functional state to another.
It is therefore important to explore biodiversity in the wild and its evolution in relation to environmental changes by using molecular methods in order to determine the diversity of cell regulatory networks that actually exist. Which networks appeared first? Is there an evolutionary pattern of molecular interaction networks? Instead of looking simply at the evolution of individual marker genes, should we also look for evolutionary patterns in network structures (combinations of interacting gene products)? We know that life evolved in a changing environment with strong discontinuities that probably channeled the existence of “possible cellular and ecological networks” (Dekel et al., 2005; Kashtan et al., 2009). The classic neo-Darwinian vision of gradual evolution by small changes and selection does not really explain (alone) the origin of variation. Population sizes, recombination, and the accumulation of neutral mutations associated with genetic drift, as well as the impact of the environment on unicellular genome evolution, are all important, albeit poorly understood factors (Colbourne et al., 2011; Fernandez and Lynch, 2011).
There is a huge source of hidden diversity in natural ecosystems that allows their robust survival. There, it is not so much the “individuals” that are important but rather the “diversity index” of an ecosystem (how many different genetic variants of a given functional type are present). This provides for adaptation to environmental changes at the level of the ecosystem as a whole, through changes in the relative abundance of more- or less-well-adapted individuals (in contrast to the black and white idea of the survival of the fittest individual species). When confronted with catastrophic changes in the environment (see, e.g., Cowen, 2000), ecosystems may change abruptly but not die completely because of this large diversity, which allows the reconstitution of different but sufficiently complex groups of organisms to form a new ecosystem.
In other words, it seems very important to look at evolution in terms of living systems embedded into … ecology. We need to think of evolution in terms of a long-term, complex self-organizing system and not just genetics and selection (see, e.g., Kauffman and Johnsen, 1991; Sole et al., 1999; Hanel et al., 2007).
AN OCEAN OF CELLULAR EVOLUTION
The fields of cell and developmental biology have been very focused around a few model systems, such as Xenopus, Drosophila, Caenorhabditis elegans, zebrafish, yeasts, and tissue culture cells (Fields and Johnston, 2005). This has proved to be extremely useful and will continue to be so to unravel fundamental molecular cell and developmental biology issues. However, this has somehow fixed the fields into a certain direction remote from the environmental constraints. Metagenomic analysis of marine samples is starting to unravel the enormous genome diversity present in the oceans (Bucklin et al., 2011; Kembel et al., 2011; Sharpton et al., 2011; Wu et al., 2011). How representative are our limited model systems of the diversity of solutions explored by evolution? How diverse are the molecular mechanisms used to generate oscillators, complex cell shapes, and metabolic networks? How are those networks affected by environmental conditions? Are they directly affected? What are the routes taken by oceanic life (bacteria, viruses, and protists) to generate the cells that first built primitive multicellular organisms (King and Carroll, 2001; King et al., 2008)? We know virtually nothing about the biodiversity of this world and do not understand the rules that govern the structure and evolution of such ecosystems. Life evolved as unicellular marine organisms exposed to severe environmental changes over a little more than the 3 billion years that preceded the emergence of metazoans 600 million years ago (Carroll, 2001; King et al., 2008). There is much to be learned from marine ecosystems about cellular evolution.
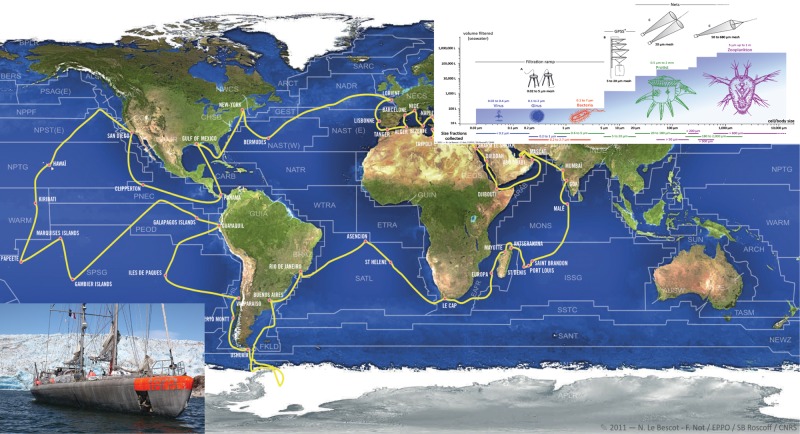
Recent around-the-world expeditions such as Tara Oceans (Figure 1; Karsenti et al., 2011) and Malaspina have collected biological samples associated with complete environmental parameters in well-defined water masses at different depths. The idea is to use quantitative imaging, metagenomics, and physicochemical oceanography to study the structure of pelagic plankton ecosystems composed of viruses, bacteria, protists, and small metazoans. This will bring back a lot of data and observations and provide food for cell biologists, modelers, and bioinformaticians to better describe the cellular origin of biodiversity, the origin of the complexity of unicellular and metazoan organisms, and the organization of ecosystems, as well as the role of environmental selection in evolution. In the oceans, microscopic ecosystems are constantly transported by currents from hot to cold regions, from poorly oxygenated to well-oxygenated areas, and from acidic to less acidic domains. Some zones of the globe become isolated from others by strong currents and temperature gradients, such as the Antarctic. Yet exchanges occur along transition zones. The oceans today are a fantastic natural laboratory of evolution, and 90% of the organisms involved are … unknown unicellular organisms! The contextual sampling of Tara Oceans associated with imaging, metagenomics, and the sequencing of individual genomes from 153 stations worldwide will provide the first set of data allowing us to explore this unknown world.
FIGURE 1:
Voyage of the Tara Oceans expedition between September 2009 and March 2012, the schooner, and the rationale of the sampling plan. The expedition crossed all major oceans except the Arctic Ocean. To characterize fully plankton ecosystems, we had to sample more than eight orders of magnitude of organism sizes. This required filtering various volumes of seawater.
It would be highly desirable for cell and developmental biologists to “lose” some precious time by enjoying the observation of the incredible organisms present in the oceans. Indeed cell biology can bring a lot to the study of evolution, just as evolution in its ecological context can bring a lot to the understanding of the self-organizational properties of cells. Besides expeditions such as Tara Oceans, Malaspina, and others, marine biology stations such as Woods Hole, Roscoff, and Villefranche, for example, should return to the fore. They should aim at promoting an interdisciplinary approach, combining cell and developmental biology with systems biology and ecology. Such a new approach would bring forward a more integrated understanding of life in the context of our planet and its long and fascinating history.
Footnotes
REFERENCES
- Athale CA, Dinarina A, Mora-Coral M, Pugieux C, Nedelec F, Karsenti E. Regulation of microtubule dynamics by reaction cascades around chromosomes. Science. 2008;322:1243–1247. doi: 10.1126/science.1161820. [DOI] [PubMed] [Google Scholar]
- Bucklin A, Steinke D, Blanco-Bercial L. DNA barcoding of marine metazoa. Annu Rev Mar Sci. 2011;3:471–508. doi: 10.1146/annurev-marine-120308-080950. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Chance and necessity: the evolution of morphological complexity and diversity. Nature. 2001;409:1102–1109. doi: 10.1038/35059227. [DOI] [PubMed] [Google Scholar]
- Caudron M, Bunt G, Bastiaens P, Karsenti E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- Chen KC, Calzone L, Csikasz-Nagy A, Cross FR, Novak B, Tyson JJ. Integrative analysis of cell cycle control in budding yeast. Mol Biol Cell. 2004;15:3841–3862. doi: 10.1091/mbc.E03-11-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke PR, Hoffman I, Draetta G, Karsenti E. Dephosphorylation of cdc25-C by a type 2A protein phosphatase: specific regulation during the cell cycle in Xenopus egg extracts. Mol Biol Cell. 1993;4:397–411. doi: 10.1091/mbc.4.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen R. History of Life. 3rd. Oxford: Blackwell Science; 2000. [Google Scholar]
- Dekel E, Mangan S, Alon U. Environmental selection of the feed-forward loop circuit in gene-regulation networks. Phys Biol. 2005;2:81–88. doi: 10.1088/1478-3975/2/2/001. [DOI] [PubMed] [Google Scholar]
- Di Ventura B, Sourjik V. Self-organized partitioning of dynamically localized proteins in bacterial cell division. Mol Syst Biol. 2011;7:457. doi: 10.1038/msb.2010.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix MA, Labbé JC, Dorée M, Hunt T, Karsenti E. Triggering of cyclin degradation in interphase extracts of amphibian eggs by cdc2 kinase. Nature. 1990;346:379–382. doi: 10.1038/346379a0. [DOI] [PubMed] [Google Scholar]
- Fernandez A, Lynch M. Non-adaptive origins of interactome complexity. Nature. 2011;474:502–505. doi: 10.1038/nature09992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE, Jr, Tsai TY, Yang Q. Modeling the cell cycle: why do certain circuits oscillate? Cell. 2011;144:874–885. doi: 10.1016/j.cell.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Fields S, Johnston M. Cell biology. Whither model organism research? Science. 2005;307:1885–1886. doi: 10.1126/science.1108872. [DOI] [PubMed] [Google Scholar]
- Fletcher DA. To model or not to model? Mol Biol Cell. 2011;22:909–910. doi: 10.1091/mbc.E11-01-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatlin JC, Bloom K. Microtubule motors in eukaryotic spindle assembly and maintenance. Semin Cell Dev Biol. 2010;21:248–254. doi: 10.1016/j.semcdb.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanel R, Kauffman SA, Thurner S. Towards a physics of evolution: critical diversity dynamics at the edges of collapse and bursts of diversification. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;76:036110. doi: 10.1103/PhysRevE.76.036110. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Hoffmann I, Clarke PR, Marcote MJ, Karsenti E, Draetta G. Phosphorylation and activation of human cdc25-C by cdc2–cyclin B and its involvement in the self-amplification of MPF at mitosis. EMBO J. 1993;12:53–63. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holy T, Leibler S. Dynamic instability of microtubules as an efficient way to search in space. Proc. Natl. Acad. Sci. USA. 1994;91:5682–5685. doi: 10.1073/pnas.91.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkovitz S, Milo R, Kashtan N, Ziv G, Alon U. Subgraphs in random networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;68:026127. doi: 10.1103/PhysRevE.68.026127. [DOI] [PubMed] [Google Scholar]
- Karsenti E. Self-organization in cell biology: a brief history. Nat Rev Mol Cell Biol. 2008;9:255–262. doi: 10.1038/nrm2357. [DOI] [PubMed] [Google Scholar]
- Karsenti E, et al. A holistic approach to marine eco-systems biology. PLoS Biol. 2011;9:e1001177. doi: 10.1371/journal.pbio.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashtan N, Parter M, Dekel E, Mayo AE, Alon U. Extinctions in heterogeneous environments and the evolution of modularity. Evolution. 2009;63:1964–1975. doi: 10.1111/j.1558-5646.2009.00684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman SA, Johnsen S. Coevolution to the edge of chaos: coupled fitness landscapes, poised states, and coevolutionary avalanches. J Theor Biol. 1991;149:467–505. doi: 10.1016/s0022-5193(05)80094-3. [DOI] [PubMed] [Google Scholar]
- Kembel SW, Eisen JA, Pollard KS, Green JL. The phylogenetic diversity of metagenomes. PLoS ONE. 2011;6:e23214. doi: 10.1371/journal.pone.0023214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Carroll SB. A receptor tyrosine kinase from choanoflagellates: molecular insights into early animal evolution. Proc Natl Acad Sci USA. 2001;98:15032–15037. doi: 10.1073/pnas.261477698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Krasinska L, Domingo-Sananes MR, Kapuy O, Parisis N, Harker B, Moorhead G, Rossignol M, Novak B, Fisher D. Protein phosphatase 2A controls the order and dynamics of cell-cycle transitions. Mol Cell. 2011;44:437–450. doi: 10.1016/j.molcel.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Loughlin R, Heald R, Nedelec F. A computational model predicts Xenopus meiotic spindle organization. J Cell Biol. 2010;191:1239–1249. doi: 10.1083/jcb.201006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Compton DA. Structural and regulatory roles of nonmotor spindle proteins. Curr Opin Cell Biol. 2008;20:101–106. doi: 10.1016/j.ceb.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Itzkovitz S, Kashtan N, Levitt R, Shen-Orr S, Ayzenshtat I, Sheffer M, Alon U. Superfamilies of evolved and designed networks. Science. 2004;303:1538–1542. doi: 10.1126/science.1089167. [DOI] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Murray AW. The cell cycle as a cdc2 cycle. Nature. 1989;342:14–15. doi: 10.1038/342014a0. [DOI] [PubMed] [Google Scholar]
- Nedelec F, Surrey T, Karsenti E. Self-organisation and forces in the microtubule cytoskeleton. Curr Opin Cell Biol. 2003;15:118–124. doi: 10.1016/s0955-0674(02)00014-5. [DOI] [PubMed] [Google Scholar]
- Nurse P. Ordering S phase and M phase in the cell cycle. Cell. 1994;79:547–550. doi: 10.1016/0092-8674(94)90539-8. [DOI] [PubMed] [Google Scholar]
- Parter M, Kashtan N, Alon U. Environmental variability and modularity of bacterial metabolic networks. BMC Evol Biol. 2007;7:169. doi: 10.1186/1471-2148-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen M, Rosenberg R, Shraiman BI, Alon U. Assigning numbers to the arrows: parameterizing a gene regulation network by using accurate expression kinetics. Proc Natl Acad Sci USA. 2002;99:10555–10560. doi: 10.1073/pnas.152046799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N, Alon U. Response delays and the structure of transcription networks. J Mol Biol. 2003;329:645–654. doi: 10.1016/s0022-2836(03)00506-0. [DOI] [PubMed] [Google Scholar]
- Sharpton TJ, Riesenfeld SJ, Kembel SW, Ladau J, O'Dwyer JP, Green JL, Eisen JA, Pollard KS. PhylOTU: a high-throughput procedure quantifies microbial community diversity and resolves novel taxa from metagenomic data. PLoS Comput Biol. 2011;7:e1001061. doi: 10.1371/journal.pcbi.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sole RV, Manrubia SC, Benton M, Kauffman S, Bak P. Criticality and scaling in evolutionary ecology. Trends Ecol Evol. 1999;14:156–160. doi: 10.1016/s0169-5347(98)01518-3. [DOI] [PubMed] [Google Scholar]
- Surrey T, Nedelec F, Leibler S, Karsenti E. Physical properties determining self-organization of motors and microtubules. Science. 2001;292:1167–1171. doi: 10.1126/science.1059758. [DOI] [PubMed] [Google Scholar]
- Tanaka TU, Desai A. Kinetochore-microtubule interactions: the means to the end. Curr Opin Cell Biol. 2008;20:53–63. doi: 10.1016/j.ceb.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth P, Lee WL, Murata T, Baskin TI. Variations on theme: spindle assembly in diverse cells. Protoplasma. 2010;248:439–446. doi: 10.1007/s00709-010-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Wollman R, Cytrynbaum EN, Jones JT, Meyer T, Scholey JM, Mogilner A. Efficient chromosome capture requires a bias in the “search-and-capture” process during mitotic-spindle assembly. Curr Biol. 2005;15:828–832. doi: 10.1016/j.cub.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Wordeman L. How kinesin motor proteins drive mitotic spindle function: lessons from molecular assays. Semin Cell Dev Biol. 2010;21:260–268. doi: 10.1016/j.semcdb.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Wu M, Halpern A, Rusch DB, Yooseph S, Frazier M, Venter JC, Eisen JA. Stalking the fourth domain in metagenomic data: searching for, discovering, and interpreting novel, deep branches in marker gene phylogenetic trees. PLoS ONE. 2011;6:e18011. doi: 10.1371/journal.pone.0018011. [DOI] [PMC free article] [PubMed] [Google Scholar]