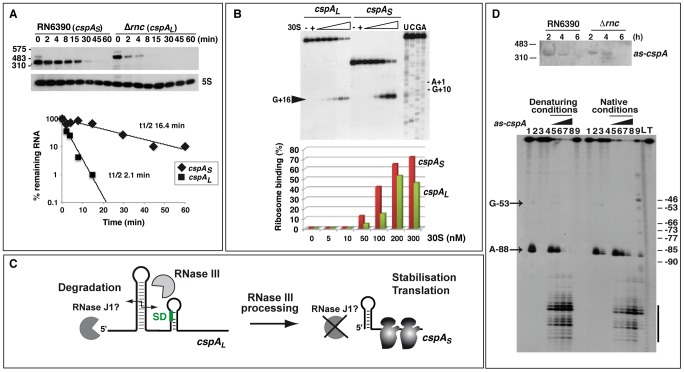
Figure 5. RNase III–dependent processing stabilizes cspA mRNA and enhances translation.
(A) Upper panel: cspA mRNA stability was assessed in RN6390 and Δrnc strains after rifampicin treatment. Expression of 5S rRNA was monitored in the same samples as a loading control. Molecular weight ladders are indicated on the left of the gel. The two forms of cspA mRNA were detected using a DIG-labeled riboprobe transcribed by T7 RNA polymerase from a PCR template amplified using the oligonucleotides 367–368 (Table S8). Lower panel: Quantification of cspA mRNA level and half-life determination in RN6390 (cspAS, diamonds) and in Δrnc strain (cspAL, squares) as a function of time. CspAL is the unprocessed mRNA and cspAS, the processed mRNA. The value corresponding to the percentage (%) of the remaining mRNA was normalized with the control experiment performed with 5S rRNA. The half-life was determined from a semi-logarithmic plot of the concentration of the mRNA over time. The slope of the best-fit line was then determined to calculate the half-life, which corresponded to the time-point where 50% of the initial mRNA amount remained. Three experiments provided reproducible results. (B) Formation of the ternary 30S initiation complex using the two forms of cspA mRNA (cspAL, and cspAS). Ternary complex formation was monitored in the presence of increasing concentrations of S. aureus 30S ribosomal subunit (5, 10, 50, 100, 200 and 300 nM), and the initiator tRNAfMet (1 µM). (−): Incubation controls without 30S. Lanes C, G, U, A: sequencing ladders of cspAL mRNA. The position of the toeprint at +16 and the +1 site corresponding to the AUG codon are indicated. Primer extension was done with the 5′ end-labeled oligonucleotide 16 (Table S8). Lower panel: Quantification of 30S ribosome binding on cspAL (green) and cspAS (red) mRNAs. Relative toeprints were calculated by relating the intensity of the band corresponding to the toeprint at +16 to the sum of the intensities of this band and the band corresponding to the full length RNA. (C) Schematic model summarizing the role of RNase III in cspA maturation. RNase III cleaves the long hairpin structure at the 5′ end of cspAL to produce an mRNA with a shorter 5′ untranslated region, which is more stable and translated with a higher efficiency. (D) Top panel: Northern blot analysis showing the expression of the antisense RNA as-cspA. Total RNAs were prepared from RN6390 (RN6390, WT) and Δrnc mutant (Δrnc) strains at 2, 4 and 6 h of growth at 37°C. Molecular weight ladders are indicated on the left of the gel. To detect as-cspA, a DIG-labeled riboprobe was transcribed in vitro with T7 RNA polymerase from a PCR template amplified with the oligonucleotides 286 and 16 (Table S8). The asRNA signal was detected after a long exposure of the autoradiography (30 min). Bottom panel: autoradiography showing the fractionation of RNase III cleavages of 5′ end-labeled cspAL mRNA alone or in the presence of the antisense RNA (as-cspA). Incubation controls of cspAL mRNA alone or with as-cspA in the absence of RNase III are shown respectively in lanes 1 and 4. The RNase III cleavage assays were done in the presence of Mg2+ (lanes 2, 5–8) or Ca2+ (lanes 3, 9) with cspAL mRNA alone (lanes 2–3) or with as-cspA (lanes 4–9). The cspA mRNA-as-cspA duplex was formed with denatured RNAs (denaturing conditions) or with RNAs, which were separately renatured (native conditions) (see Text S1). Increasing concentrations of asRNA were used: 10 nM (lane 5), 25 nM (lane 6), 50 nM (lane 7), and 100 nM (lanes 4, 8, 9). Lanes L, T: alkaline ladder and RNase T1 performed on cspAL mRNA under denaturing conditions, respectively. Lanes 3, 9 (native conditions): the experiments performed in the presence of Ca2+ under native conditions show a residual activity of RNase III due to the presence of Mg2+, which was used to fold the RNAs prior to complex formation. The arrow indicates the RNase III cleavage at position A-88 occurring in free cspAL mRNA, and the bar shows the strongest cleavages induced by the as-cspA binding.