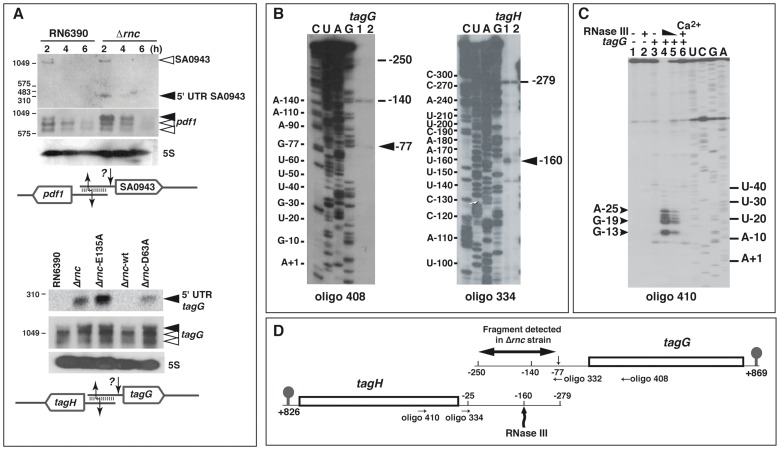
Figure 8. RNase III cleaves mRNAs with overlapping 5′ untranslated regions (UTR).
(A) Effect of RNase III on the overlapping 5′ UTRs of SA0943/pdf1 (top) and tagG/tagH (bottom). RNA levels were monitored on Northern blots in various strains: RN6390 (WT), Δrnc, and Δrnc transformed with plasmids expressing E135A RNase III mutant (Δrnc-E135A), the wild type RNase III (Δrnc-wt), or the D63A RNase III mutant (Δrnc-D63A). DIG labeled riboprobes were used to monitor the expression of mRNAs. The riboprobes were transcribed in vitro with T7 RNA polymerase using PCR templates amplified with the oligonucleotides 426/427 (SA0943), 429/433 (pdf1), 333/334 (5′UTR tagG) and 392/393 (tagG) (Table S8). Schematic drawings summarizing the data are given below the Northern blot experiments. They show the overlapping 5′UTRs that form a typical RNase III binding substrate. A second cleavage from an unknown RNase (?) generated RNA fragments of 300 nts corresponding to the size of the 5′UTRs of SA0943 or tagG, which only accumulated in the Δrnc strain. (B) Primer extension analysis was performed on total RNAs prepared from exponential phase of growth. Oligonucleotides 408 and 334 were used to probe the 5′ ends of tagG and tagH mRNAs, respectively. Lanes 1, 2: samples prepared from RN6390 and Δrnc mutant strains, respectively. The 5′ ends (−140 in tagG mRNA and −25 in tagH mRNA) were detected both by primer extension and by RACE experiments performed on circularized RNAs (Table S6). Additional 5′ ends as well as putative RNase cleavage sites are indicated. The nucleotides are numbered relatively to the AUG start codon. Lanes C, U, A, G: sequencing ladders performed on tagG and tagH mRNAs, which were transcribed from PCR templates amplified with primers 331/221 and 333/384, respectively. (C) RNase III cleavage assays performed with in vitro transcribed tagH mRNA. Cleavage sites were assigned by primer extension using the 5′ end-labeled oligonucleotide 410. Controls were done with free mRNA (lane 1) or bound to tagG mRNA (lane 3); RNase III cleavage assays were performed on the mRNA alone (lane 2) or bound to tagG mRNA in a buffer containing Mg2+ (lanes 4, 5) or Ca2+ (lane 6). Reactions were set with 0.6 µM (lane 2, 5, 6) and 0.8 µM (lane 4) of wild-type RNase III; lanes U, C, G, A: sequencing ladders. (D) Schematic representation of the tagG-tagH locus. The RNase III cleavage site in the 5′UTR of tagH mRNA at position −160 is shown. An additional cleavage site induced by an unknown RNase is designated by a small arrow at position −77 of tagG mRNA. The fragment detected by Northern blot in Δrnc mutant strain (Figure 8A, lower panel) and the hybridization sites of the primers are indicated. The 3′ ends of both mRNAs were identified by RACE on circularized mRNAs (+826 for tagH and +869 for tagG, respectively).