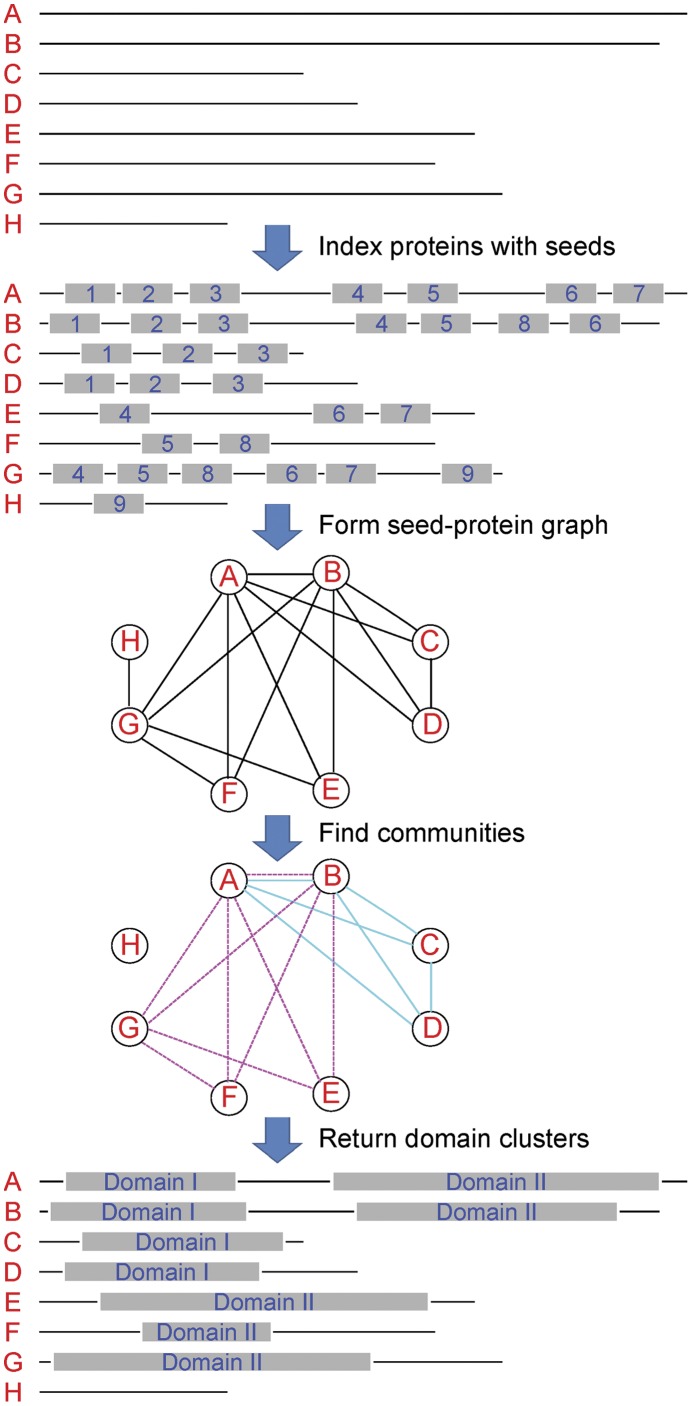
Figure 1. Outline of SECOM.
Given a set of protein sequences (“A” to “H”), SECOM first finds all the hash seeds (“1” to “9”) that appear at least twice in this set. A seed-protein graph is then built, in which each node is a protein sequence and two nodes are connected if they share at least one hash seed. The highly connected subgraphs (i.e., communities) are found in this graph. The communities can be overlapping and each of them (“Domain I” and “Domain II”) is a predicted domain cluster by SECOM.