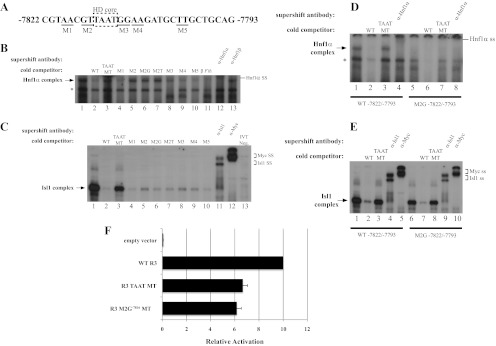
Fig. 3.
Hnf1α activates R3 through the bp −7816/−7811 element. A, The location of the M1 through M5 mutations within bp −7822/−7793 probe is illustrated, with the homeodomain (HD) core motif enclosed by the dashed box. B, The M2, M2G−7816, M2T−7815, and TAAT mutants failed to compete at a 50-fold molar excess for Hnf1α binding in βTC3 nuclear extracts, whereas M1, M3, M4, and M5 were effective competitors. C, In vitro translated Isl1-myc binding was reduced in the presence of 50-fold excess of the M2, M3, and M4 competitors, although less efficiently than wild type (WT), M2G−7816, M2T−7815, and M5. Isl1 and myc-epitope antibodies were used to localize the Isl1 binding complex, which was not detected with the in vitro translation reactions performed with the pcDNA3.1 vector (termed IVT neg). D and E, The bp −7822/−7793 M2G−7816 probe binds Isl1, but not Hnf1α. WT or M2G−7816 bp −7822/−7793 probes were incubated with βTC-3 nuclear extract (D) or Isl1-myc protein (E), WT or core TAAT MT competitor, and Hnf1α-, Isl1-, or myc-specific antibodies. F, The Hnf1α binding defective M2G−7816 mutation reduced transfected R3-driven reporter activity in βTC-3 cells. The reduced activity of M2G−7816 was similar to the TAAT mutant, which was deficient in both EMSA Hnf1α and Isl1 binding activity. *, Nonspecific complex; ss, supershift.