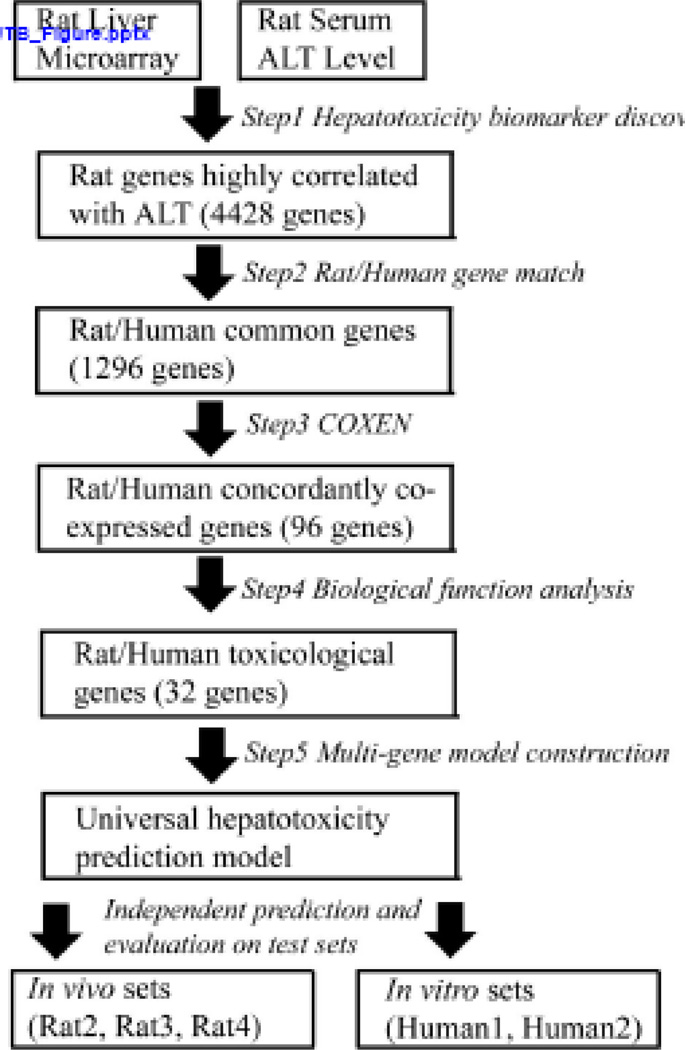
Figure 1. Schematic diagram of the computational model construction and validation processes.
Our hepatocellular toxicity prediction was developed by five distinct steps - the first was the identification of hepatocellular toxicity biomarker genes by correlation analysis with serum ALT. The second was the homologous gene match between human and rat while the third was the selection of the COXEN biomarkers among human-homologous hepatocellular toxic biomarkers obtained from the second step. In the fourth step, we further selected the biomarkers known to be directly related to liver toxicity and cell death performed by Ingenuity Pathway Analysis. This functional analysis step led to the final 32 biomarkers of hepatocellular toxicity. The last step was the multivariate prediction model construction based on the final 32 genes.