Abstract
Altered lipid metabolism underlies several major human diseases, including obesity and type 2 diabetes. However, lipid metabolism pathophysiology remains poorly understood at the molecular level. Insulin is the primary stimulator of hepatic lipogenesis through activation of the SREBP-1c transcription factor. Here we identified cyclin-dependent kinase 8 (CDK8) and its regulatory partner cyclin C (CycC) as negative regulators of the lipogenic pathway in Drosophila, mammalian hepatocytes, and mouse liver. The inhibitory effect of CDK8 and CycC on de novo lipogenesis was mediated through CDK8 phosphorylation of nuclear SREBP-1c at a conserved threonine residue. Phosphorylation by CDK8 enhanced SREBP-1c ubiquitination and protein degradation. Importantly, consistent with the physiologic regulation of lipid biosynthesis, CDK8 and CycC proteins were rapidly downregulated by feeding and insulin, resulting in decreased SREBP-1c phosphorylation. Moreover, overexpression of CycC efficiently suppressed insulin and feeding–induced lipogenic gene expression. Taken together, these results demonstrate that CDK8 and CycC function as evolutionarily conserved components of the insulin signaling pathway in regulating lipid homeostasis.
Introduction
Dysregulation of lipid metabolism is closely associated with major human diseases such as obesity, type 2 diabetes, and cardiovascular disease (1–4). However, the physiologic and pathophysiologic regulation of lipid metabolism is a highly complex and integrative process that remains poorly understood at the molecular level. Among the known lipogenic regulators, the SREBP transcription factors are pivotal activators of key enzymes responsible for hepatic biosynthesis of fatty acids and cholesterol (5–8) and play an important role in the development of fatty liver and dyslipidemia (9).
The three mammalian SREBP transcription factors, SREBP-1a, -1c, and -2, are synthesized as inactive precursors that are tethered to the ER membrane (10). Reduction of intracellular sterols results in the transportation of SREBP-2 to the Golgi, where it undergoes proteolytic maturation. Then, the N-terminal fragment of SREBP-2 translocates into the nucleus and activates transcription of target genes (11). Similarly, the two SREBP-1 isoforms, SREBP-1a and -1c, are also processed in the Golgi to generate the mature SREBP-1 protein (12, 13). Unlike SREBP-2, SREBP-1c is primarily activated by insulin (14). Both SREBP-1a and SREBP-1c proteins are produced by the same gene, SREBF1, by two distinct promoters and alternative splicing (15). Their amino acid sequences differ only at the very N-terminal end. The unique region of SREBP-1a is part of its transactivation domain (16). SREBP-1a and SREBP-1c have different expression profiles: SREBP-1a is highly expressed in proliferating cells, such as cancer cells, while SREBP-1c is the predominant form in normal cells, particularly hepatocytes (17).
CDK8 and its regulatory partner CycC have been reported as being subunits of the Mediator complexes in mammalian cells (18, 19). The mammalian Mediators are large protein complexes containing up to 30 distinct subunits, depending on starting materials and biochemical purification protocols, and play a critical role in bridging the signals from transcription factors to the basal transcription apparatus (20–22). Biochemical purifications have identified at least two groups of Mediator complexes, the small Mediator and the large Mediator, with the latter containing an extra submodule of CDK8, CycC, MED12, and MED13 (19). In chromatin-based in vitro transcription assays, the small Mediator can activate gene transcription, while the large Mediator is inactive (19). Interestingly, recent studies have established a role of CDK8-CycC in tumorigenesis (20). However, the in vivo functions and regulation of CDK8 and CycC are still poorly understood.
In this study, we identify CDK8 and CycC as key repressors for lipogenic gene expression, de novo lipogenesis, and lipid accumulation in Drosophila and mammals. This function of CDK8-CycC occurs through site-specific phosphorylation on nuclear SREBP-1c protein, resulting in rapid degradation of this central regulator of lipid metabolism. Since CDK8 and CycC are reduced upon feeding or by insulin, concomitant with increased levels of nuclear SREBP-1c protein, the CDK8-CycC complex appears to function downstream of the insulin signaling as a key regulator of de novo lipogenesis.
Results
CDK8 and CycC regulate lipid levels and lipogenic gene expression in Drosophila.
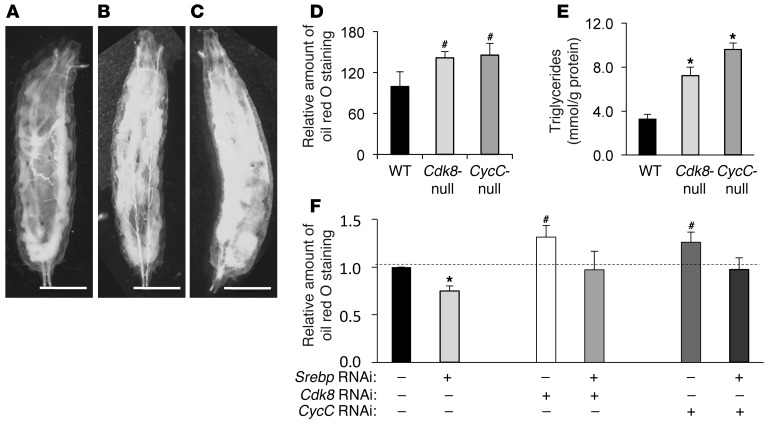
CDK8 and CycC are highly conserved in eukaryotes; thus, we first used Drosophila as a model organism to study function and regulation of CDK8 and CycC in vivo. Compared with wild-type (Figure 1A), Cdk8-null (Figure 1B) and CycC-null (Figure 1C) larvae are less transparent, suggesting that the mutant larvae possess more fat tissue. Intrigued by this observation, we stained fat body of larvae, which is the predominant tissue for regulating lipid homeostasis in Drosophila, with oil red O for neutral lipids (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI61462DS1) and extracted the dye with isopropanol. As shown in Figure 1D, we detected significantly more oil red O staining in Cdk8- and CycC-null larvae. Quantitative measurements of triglycerides in Cdk8- and CycC-null larvae further confirmed the significant increase in neutral lipids as compared with control (Figure 1E). To avoid potential nonspecific effects of Cdk8 or CycC mutations on lipid metabolism during development, we generated transgenic RNAi lines that allow tissue-specific knockdown of CDK8 or CycC. Similarly, we observed a significant increase in lipid levels in larvae with either CDK8 or CycC knockdown by RNAi whose expression is driven by fat body–specific FB-Gal4 (Figure 1F) or Adh-Gal4 (data not shown). Taken together, these results suggest that CDK8 and CycC are required for inhibition of lipid accumulation in Drosophila larvae.
Figure 1. Role of CDK8 and CycC in lipid accumulation in fat bodies of Drosophila larvae.
Representative light micrographic images for larvae (scale bars: 1.0 mm) of (A) wild-type (w1118), (B) Cdk8-null (k185), and (C) CycC-null (y5) Drosophila. (D) Quantitative measurement of oil red O staining in Drosophila larvae of the indicated genotypes. (E) Triglyceride levels in Drosophila larvae of the indicated genotypes. (F) Quantitative measurement of oil red O staining in Drosophila larvae with fat body–specific expression of the indicated RNAi; the background control was FB-Gal4. Data represent mean ± SD of 15 larvae per genotypes for oil red O staining or 3 random pools of the same genotype for triglyceride measurement. *P < 0.01, #P < 0.001 versus control. Data are representative of independent experiments repeated at least 3 times.
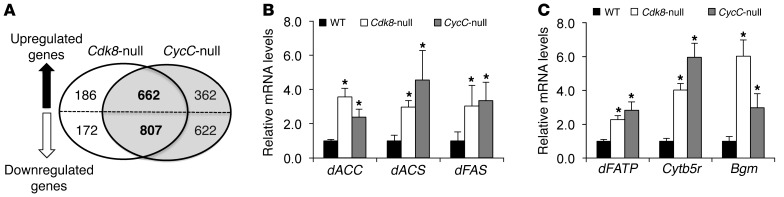
To investigate the underlying mechanisms of increased lipid levels in Cdk8 and CycC mutants in an unbiased fashion, we examined the global gene expression profiles in homozygous null mutant larvae of Cdk8 and CycC at the late third instar stage by cDNA microarray analysis. Both Cdk8K185 and CycCY5 are null alleles, and homozygous mutants are lethal as pupae (23). Compared with the control, a number of genes in Cdk8-null mutants were significantly altered, with a cutoff at 1.5-fold and a false discovery rate of less than 0.05 (Figure 2A). More than 80% of these genes (662 of upregulated and 807 of downregulated genes) were also altered in CycC-null mutants in the same fashion (Figure 2A). The high percentage of overlapping genes supports a model whereby CycC is required for activation of CDK8 (20, 23–26). In addition, the known dE2F1 target genes were also found to be upregulated in this microarray analysis (data not shown), confirming our previous finding that CDK8 inhibits E2F1-dependent transcription (27).
Figure 2. Role of CDK8 and CycC in gene expression in Drosophila larvae.
(A) Number of genes whose expression was significantly changed in Cdk8-null and CycC-null Drosophila larvae identified by cDNA microarray analysis. (B and C) The mRNA levels of representative lipogenic genes determined by qRT-PCR using rp49 as the invariant control. Data represent mean ± SD of 3 random pools of the same genotype. *P < 0.01 versus control.
Consistent with our observation of the phenotypic changes in Cdk8- or CycC-null larvae, many genes involved in lipid metabolism were significantly upregulated in those mutants (Supplemental Table 1). These genes include fatty acid synthase (dFAS), acetyl-CoA carboxylase (dACC), and acetyl-CoA synthetase (dACS), which are important enzymes for fatty acid biosynthesis. As confirmed by reverse transcription and quantitative PCR (qRT-PCR), we observed a significant increase in transcripts of dACC, dACS, dFAS, and other genes in Cdk8- and CycC-null mutants compared with the control (Figure 2, B and C), suggesting that CDK8 and CycC are required for inhibition of the transcription of these genes.
Interestingly, the mammalian homologs of several genes in Supplemental Table 1 are known transcriptional targets of SREBP transcription factors (28, 29), which are the master regulators of lipid homeostasis in diverse organisms (10, 30, 31). Consistent with the lipogenic function of SREBP, knocking down Drosophila SREBP (dSREBP) in fat body significantly reduced lipid levels (Figure 1F). Thus, we postulate that CDK8 and CycC may regulate the expression of lipogenic genes through repression of SREBP-mediated transcription. To test this hypothesis, we knocked down both CDK8 (or CycC) and dSREBP in fat body and observed a significant reduction in neutral lipid levels compared with knockdown of CDK8 (or CycC) alone (Figure 1F). These results suggest that the effect of CDK8 or CycC knockdown on lipid accumulation is likely dependent on SREBP. However, the mRNA levels of dSREBP and dSCAP were not significantly changed in Cdk8- and CycC-null mutants (Supplemental Table 1), suggesting that the effect of CDK8 and CycC on SREBP target gene expression in Drosophila was less likely mediated through regulation of the transcription or maturation of dSREBP itself.
CDK8 and CycC repress the lipogenic program in mammalian hepatocytes.
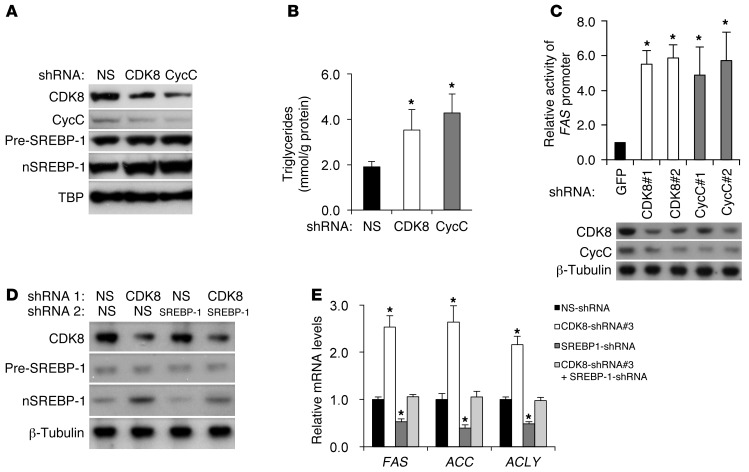
To determine whether CDK8 regulation of lipid accumulation and lipogenic gene expression is conserved in mammalian cells, we knocked down CDK8 or CycC in primary culture of rat hepatocytes using lentiviral shRNA (Figure 3A). Knockdown of CDK8 or CycC resulted in a significant increase in triglycerides (Figure 3B), suggesting that as in Drosophila larvae, CDK8 and CycC are required for inhibition of intracellular triglyceride accumulation in primary rat hepatocytes. To test whether CDK8 and CycC also regulate the transcription of FAS, we knocked down CDK8 or CycC in HepG2 cells that were cotransfected with the FAS luciferase reporter (32). As shown in Figure 3C, CDK8 or CycC knockdown with two independent shRNAs, as we used previously (27), resulted in significant elevation of FAS promoter activity, indicating that CDK8 and CycC negatively regulate the FAS promoter. Consistent with the concept that a functional interaction of CDK8 and CycC is necessary for stability of the CDK8-CycC complex, CDK8 knockdown in mammalian cells resulted in a concomitant reduction in CycC, while CycC knockdown caused a parallel reduction in CDK8 in cultured cells (Figure 3, A and C). In addition, CDK8 or CycC knockdown (Figure 3D) significantly increased the mRNA levels of endogenous SREBP target genes, such as FAS, ACC, ATP-citrate lyase (ACLY), and stearoyl CoA desaturase (SCD1), in three hepatocyte lines: primary rat hepatocytes (Figure 3E), FAO cells (data not shown), and HepG2 cells (Supplemental Figure 2A). The effect of CDK8 or CycC knockdown was compromised when SREBP-1 was co-depleted in those cells by shRNA (Figure 3E and Supplemental Figure 2A), similar to what we observed in Drosophila (Figure 1F). These results support the hypothesis that regulation of lipogenic gene expression by CDK8 is dependent on SREBP-1. Furthermore, CDK8 knockdown significantly stimulated the activity of a promoter with wild-type SREBP-1 binding sites (SRE) in HEK293 cells, but had no effects when the SRE site was inactivated by point mutations (Supplemental Figure 2B), further suggesting that CDK8 represses SREBP-1–dependent gene expression. Consistent with the mRNA level and promoter activity of FAS gene, FAS protein levels in primary rat hepatocytes (data not shown) and HepG2 cells (Figure 4A) were also significantly elevated when CDK8 or CycC was knocked down. Taken together, our data suggest that CDK8 and CycC regulate SREBP-dependent gene expression in mammalian hepatocytes.
Figure 3. CDK8 and CycC regulate SREBP-1–dependent lipogenic gene expression in mammalian hepatocytes.
(A and D) Immunoblots for the indicated proteins in primary rat hepatocytes after treatment with lentivirus expressing the indicated shRNA. Pre-SREBP-1, precursor SREBP-1; nSREBP-1, nuclear SREBP-1. (B) Effects of CDK8 or CycC knockdown by lentiviral shRNA on triglyceride levels in isolated primary rat hepatocytes. (C) Effect of CDK8 or CycC knockdown by shRNA on FAS promoter–driven firefly luciferase mRNA levels in HepG2 cells as measured by qRT-PCR. The β-actin–driven Renilla luciferase served as control. (E) mRNA levels of endogenous SREBP target genes in primary rat hepatocytes were detected by qRT-PCR after treatments of the indicated lentiviral shRNA. Cyclophilin B served as the invariant control, and the mRNA levels of each gene were normalized to those in the non-silencing (NS) shRNA–treated sample. Data represent mean ± SD of 3 independent treatments. *P < 0.01 versus control.
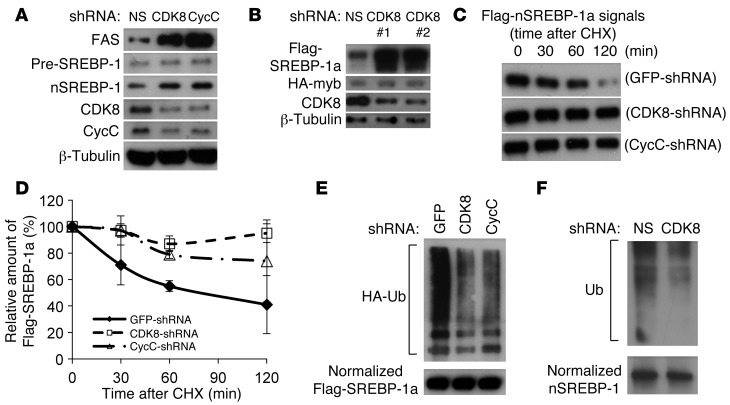
Figure 4. CDK8 and CycC regulate SREBP-1 protein stability.
(A) Effects of CDK8 and CycC knockdown on protein levels of FAS and SREBP-1 in HepG2 cells. (B) Effects of CDK8 knockdown on protein levels of overexpressed Flag-tagged nuclear SREBP-1a in HEK293 cells. The HA-tagged fusion protein of the transactivation domain of Myb and Gal4 DNA-binding domain served as control. (C) Representative immunoblots and (D) relative levels of overexpressed Flag-tagged nuclear SREBP-1a in HEK293 cells in the presence of cycloheximide (CHX) for the indicated amount of time after CDK8 or CycC knockdown. (E) Effects of CDK8 or CycC knockdown on SREBP-1a ubiquitination. Flag-tagged nuclear SREBP-1a was cotransfected with HA-tagged ubiquitin (HA-Ub) and the indicated shRNA plasmids in HEK293 cells. After approximately 40 hours of culture followed by 3 hours of treatment with 0.1 mM MG132, cell lysates were prepared. The amount of SREBP-1a proteins was normalized after immunoprecipitation from cell lysate, and the presence of HA-ubiquitin modifications was detected by immunoblotting using anti-HA antibody. (F) Endogenous nuclear SREBP-1 proteins were immunoprecipitated from HEK293 cells after lentiviral CDK8 (or NS) shRNA treatment for 48 hours, followed by 3 hours of treatment with 0.1 mM MG132. The presence of ubiquitinated proteins was detected by anti-ubiquitin antibody.
Cdk8 and CycC are involved in regulating nuclear SREBP-1 protein stability.
To elucidate the molecular mechanisms of CDK8 inhibition of SREBP-1–mediated lipogenic gene expression, we first asked whether CDK8 affects the abundance of nuclear SREBP-1 in mammalian cells. CDK8 or CycC knockdown resulted in a significant increase in endogenous nuclear SREBP-1 protein in primary rat hepatocytes (Figure 3A) and HepG2 cells (Figure 4A), with no significant change in the protein levels of the SREBP-1 precursor (Figure 3A and Figure 4A), suggesting that CDK8 regulates the abundance of the nuclear form of this transcription factor.
The nuclear/active form of SREBP-1 is processed from its precursor initially located in ER membrane (13). Since CDK8 is predominantly a nuclear protein (33), it is less likely that CDK8 can directly regulate the proteolytic maturation process of SREBP-1 in ER or Golgi apparatus, which is also supported by our data in primary rat hepatocytes (Figure 3A and Figure 4A). Thus, to further analyze the effect of CDK8 on nuclear SREBP-1 protein levels, we initially overexpressed a Flag-tagged nuclear form of SREBP-1a in HEK293 cells by transient transfection. As a control, an HA-tagged fusion protein of the Gal4 DNA-binding domain and Myb transactivation domain, which is neither physically nor functionally associated with CDK8 or CycC (32, 34), was coexpressed. As shown in Figure 4B, depletion of CDK8 by shRNAs caused accumulation of the nuclear form of SREBP-1a. Similar results were obtained when CycC was knocked down (data not shown). The increased accumulation of the nuclear form of SREBP-1a in HEK293 cells caused by CDK8 or CycC knockdown was due to decreased protein degradation as assayed by Western blots in the presence of cycloheximide, an inhibitor of protein synthesis (Figure 4, C and D). These data support a model in which CDK8-CycC regulates SREBP target gene expression at least in part by controlling the stability of nuclear SREBP-1 proteins.
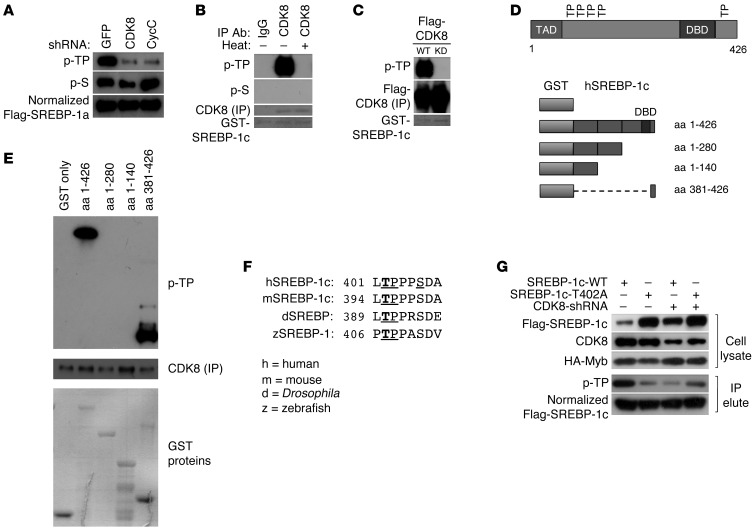
CDK8 directly phosphorylates SREBP-1.
Since ubiquitination is required for SREBP-1 degradation (35), we determined whether CDK8 and CycC affected SREBP-1a ubiquitination by coexpressing HA-tagged ubiquitin with Flag-tagged nuclear SREBP-1a in HEK293 cells. As shown in Figure 4E, the amount of ubiquitinated SREBP-1a was significantly decreased in cells with CDK8 or CycC knockdown by shRNAs. Moreover, CDK8 knockdown also decreased the ubiquitination levels of endogenous SREBP-1 (Figure 4F).
Phosphorylation of SREBPs has been shown to facilitate their binding to the E3 ligase SCFFbw7b, thus controlling their ubiquitination and subsequent degradation (35). Because CDK8 can directly phosphorylate several proteins in multicellular organisms (27, 36–38), we tested the possibility that CDK8 alters SREBP-1a stability through direct phosphorylation. Since the consensus sequence motifs for cyclin-dependent kinases are generally threonine (T) or serine (S) followed by a proline residue (TP or SP), we used a commercially available antibody that specifically recognizes phosphorylated TP (p-TP) as well as an antibody that can recognize phosphorylated serine (p-S) to detect the phosphorylation status of SREBP-1a. By Western blot analysis of immunoprecipitated SREBP-1a, we found that CDK8 or CycC knockdown in HEK293 cells dramatically decreased the phosphorylation levels of SREBP-1a at TP motifs, but had no significant effects on phosphorylation at serine residues (Figure 5A), suggesting that CDK8 or CycC only regulates threonine phosphorylation of SREBP-1a. To determine whether CDK8 can also directly phosphorylate SREBP-1c, we performed in vitro kinase assays in which endogenous CDK8 proteins were immunopurified from nuclear extracts of HEK293 cells, and recombinant GST–SREBP-1c (amino acids 1–426) protein was used as the potential substrate. As shown in Figure 5B, native CDK8, but not heat-inactivated CDK8, could phosphorylate SREBP-1c in vitro at TP motifs, but not serine residues. To eliminate the possibility of contamination by other cellular kinases, we immunopurified Flag-tagged CDK8 proteins from nuclear extracts of HEK293 cells that were transfected with either wild-type or a kinase-dead mutant of CDK8. As shown in Figure 5C, wild-type CDK8, but not the kinase-defective mutant, could phosphorylate SREBP-1c in vitro, confirming that SREBP-1c is a direct substrate of CDK8. Moreover, only CDK8, but not the closely related nuclear CDK7 and CDK9, was able to phosphorylate SREBP-1c at TP motifs, and none of them was able to phosphorylate SREBP-1c on serine residues in vitro (data not shown). Consistent with a previous report (39), all of these kinases could phosphorylate the CTD tail of RNA polymerase II (RNAPII) in vitro (data not shown).
Figure 5. Direct phosphorylation of SREBP-1 by CDK8.
(A) Effects of CDK8 or CycC knockdown on SREBP-1a phosphorylation in HEK293 cells. Flag-tagged nuclear SREBP-1a was cotransfected with the indicated shRNA plasmids in HEK293 cells. Cell lysates were prepared after approximately 40 hours of culture. SREBP-1a proteins were immunoprecipitated from cell lysate and normalized. The presence of p-TP motifs and p-S residues was detected by immunoblotting using specific antibodies. (B and C) In vitro kinase assays were performed to determine kinase activity toward GST-SREBP-1c (aa 1–426) at TP motifs using the following immunopurified kinases from nuclear extract: (B) endogenous CDK8; and (C) Flag-tagged wild-type or kinase-dead (KD) mutant CDK8 by transient transfection. (D) SREBP-1c fragments used for mapping CDK8 target sites. (E) Identification of CDK8 target site by in vitro kinase assays. (F) Conservation of CDK8 phosphorylation site in SREBPs. (G) Effects of CDK8 knockdown on Flag-tagged WT or T402A mutant SREBP-1c protein levels and phosphorylation at TP motifs in HEK293 cells.
There are 5 TP motifs within the nuclear form of human SREBP-1c (Figure 5D). To determine which TP motifs are targeted by CDK8, we performed in vitro kinase assays with several truncated forms of SREBP-1c (Figure 5D). Only the fragments containing the T402 residue (Figure 5E), which is located in a highly conserved region (Figure 5F), can be phosphorylated by CDK8 in vitro, suggesting that the T402 residue of human SREBP-1c is likely the direct target site of CDK8. To determine whether CDK8 affects phosphorylation of SREBP-1c T402 in cultured cells, we mutated this residue to alanine (T402A mutation) and transfected the Flag-tagged mutant as well as wild-type SREBP-1c into HEK293 cells. Consistent with our in vitro results, T402A mutant SREBP-1c was more stable than the wild type SREBP-1c in HEK293 cells (Figure 5G). Similar to our data on SREBP-1a (Figure 4B), CDK8 knockdown also induced accumulation of nuclear SREBP-1c, but the protein levels of the T402A mutant, unlike wild-type SREBP-1c, were not affected by CDK8 knockdown (Figure 5G). In addition, the phosphorylation levels of TP motifs in T402A mutant SREBP-1c were dramatically decreased, albeit not completely lost (Figure 5G), and CDK8 knockdown no longer had significant effects on TP phosphorylation of T402A mutant SREBP-1c (Figure 5G). These results strongly support the hypothesis that the conserved T402 is the primary target of TP motif phosphorylation by CDK8, but the non-conserved TP motifs can have minor phosphorylation by other unknown kinases in HEK293 cells. Functionally, CDK8 knockdown can stimulate wild-type SREBP-1c– but not T402A SREBP-1c–mediated activation of the FAS promoter (Supplemental Figure 3). Our results are consistent with a previous report showing that phosphorylation of the analogous T426 residue of SREBP-1a is critical for its binding to the E3 ligase SCFFbw7b, which is also involved in regulating nuclear SREBP-1a protein stability (35). Thus, our data directly demonstrate that CDK8 functions as a novel kinase of SREBP-1c.
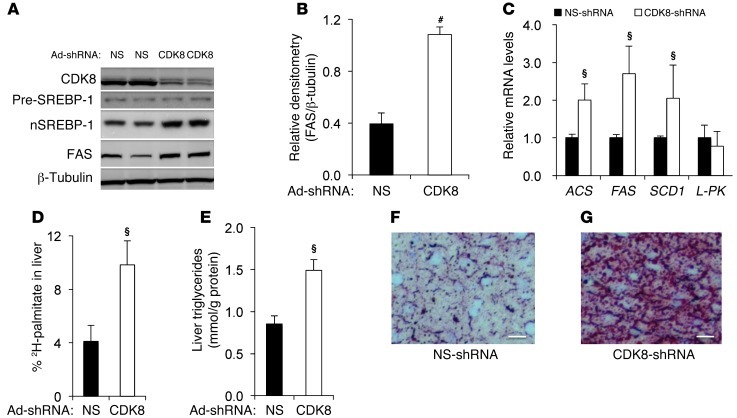
Knockdown of CDK8 causes lipid accumulation in mice.
The regulation of lipogenesis in cultured hepatocytes, particularly cancer cell lines, may not necessarily reflect the physiologic regulation in vivo. However, conventional CDK8 knockouts are embryonic lethal (40), and conditional CDK8-knockout mice are not yet available. Thus, to avoid the developmental defects of Cdk8 gene ablation, we transiently knocked down CDK8 in mouse livers by tail vein injection of adenovirus expressing specific shRNA against CDK8 (Figure 6A). Similar to our data from cell culture, CDK8 knockdown in mouse liver caused a significant increase in protein levels of FAS (Figure 6, A and B) and the nuclear, but not precursor, form of SREBP-1c (Figure 6A). In addition, we found that the SREBP-1c target genes FAS, ACS, and SCD1 were all significantly upregulated in CDK8-knockdown mouse livers compared with non-silencing shRNA–adenovirus infected samples (Figure 6C). In addition, CDK8 knockdown in liver had no significant effect on Srebp1 or Srebp2 mRNA expression (Supplemental Figure 4A) or L-pyruvate kinase (Figure 6C). The latter is not regulated by SREBP-1c (41), but it is a target of another important lipogenic transcription factor, carbohydrate regulatory element binding protein (ChREBP) (42, 43). These results suggest an inhibitory role of CDK8 on lipogenic gene expression in vivo through the regulation of nuclear SREBP-1c. Consistent with the increased lipogenic gene expression, the hepatic de novo synthesis rate of palmitate was significantly elevated by injection of adenoviral CDK8 shRNA (Figure 6D). As a result, hepatic triglyceride levels were also significantly increased, by about 2-fold (Figure 6E), with accumulation of neutral lipids in mouse livers as detected by oil red O staining (Figure 6, G versus F), revealing a phenotype of early-stage fatty liver in CDK8-knockdown mice. Strikingly, CDK8 knockdown also resulted in a concomitant increase in plasma triglycerides (~5-fold) (Supplemental Figure 4B), but no change in circulating insulin (Supplemental Figure 4C), free fatty acids (Supplemental Figure 4D), or glycerol (Supplemental Figure 4E). These results suggest that hypertriglyceridemia in CDK8-knockdown mice was not due to changes in adipocyte lipolysis. Thus, our data from mouse livers support the hypothesis that CDK8 plays a critical role in regulating de novo lipogenesis and triglyceride levels in mammals.
Figure 6. CDK8 knockdown in mouse liver increases de novo lipogenesis.
(A) Representative immunoblots for the indicated proteins after tail vein injection of adenoviruses expressing shRNA in mouse liver. (B) Relative amount of FAS proteins after the indicted treatments. (C–G) Effects of CDK8 knockdown by adenoviral shRNA in mouse livers in vivo on lipogenic gene expression as detected by qRT-PCR (C), synthesis rate of hepatic palmitate as analyzed by GC-MS (D), hepatic triglyceride levels (E), and hepatic lipid accumulation as visualized by oil red O staining (G; control is shown in F; scale bar: 0.01 mm). L-PK, L-pyruvate kinase. Data represent mean ± SD (n = 6 mice for each treatment). For CDK8 shRNA, §P < 0.05 and #P < 0.001 versus control.
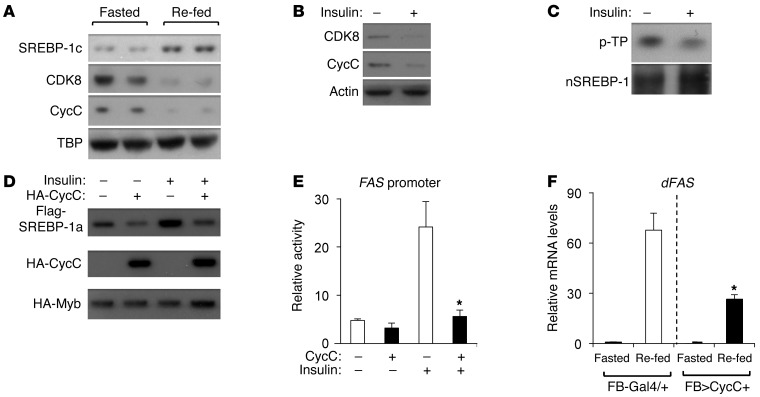
Regulation of CDK8 and CycC by nutrients.
Since food intake generally stimulates lipogenesis in liver, we examined the protein levels of nuclear SREBP-1c, CycC, and CDK8 in mouse liver in fasted and re-fed states. Consistent with a previous report (44), compared with mice fasted for 12 hours, those re-fed for 5 hours had higher levels of nuclear SREBP-1c (Figure 7A). Interestingly, re-feeding resulted in a significant decrease in nuclear levels of both CDK8 and CycC proteins in mouse liver (Figure 7A), suggesting that CDK8 and CycC are regulated during fasting and re-feeding. Although a complicated set of changes takes place from the fasting to fed state, insulin is a primary lipogenic factor upon food intake, and it stimulates de novo lipogenesis primarily by stimulating SREBP-1c at the levels of transcription, processing, and nuclear protein stability (14). Thus, we tested whether insulin has any effects on CDK8 or CycC levels in hepatocytes. As shown in Figure 7B, insulin treatment caused downregulation of both CDK8 and CycC proteins in primary rat hepatocytes. We also observed a similar effect of insulin in FAO and HEK293 cells (data not shown). Moreover, insulin treatment also markedly decreased the phosphorylation of nuclear form of endogenous SREBP-1c at TP sites in primary rat hepatocytes (Figure 7C).
Figure 7. Regulation of CDK8 and CycC by feeding and insulin.
(A) The indicated nuclear protein levels in livers of mice that were fasted for 12 hours (Fasted) or fasted for 12 hours followed by 5 hours re-feeding with normal mouse chow (Re-fed). (B and C) Effects of 200 nM insulin on the protein levels of CDK8 and CycC (B) and phosphorylation levels at TP motifs of immunoprecipitated SREBP-1c (C) in primary rat hepatocytes for 1 hour. (D) Effects of overexpressed CycC on insulin-mediated regulation of nuclear SREBP-1a in HEK293 cells. HA-tagged Gal4 DNA-binding domain–fused Myb-TAD served as the invariant control. (E) Effects of overexpressed CycC on insulin-induced activation of the FAS promoter in HEK293 cells by luciferase reporter assays. The firefly luciferase activity of each sample was normalized by the Renilla luciferase activity of cotransfected reporter under the control of a basal promoter (n = 3). (F) In Drosophila larvae, overexpression of CycC in fat body (FB-Gal4/UAS-CycC [FB>CycC+]) inhibits the re-feeding–induced increase in lipogenic gene expression. The third instar larvae were either fasted for 15 hours only or fasted for 15 hours followed by re-feeding for 5 hours. n = 3 with 10 larvae in each group; *P < 0.01 versus control (in the presence of insulin or re-fed).
After establishing the role of feeding and insulin in downregulating CDK8-CycC, we examined whether overexpression of CycC and/or CDK8 can antagonize the effects of insulin. As expected, insulin treatment caused accumulation of overexpressed Flag-tagged nuclear form of SREBP-1a in HEK293 cells (Figure 7D, lane 3 vs. lane 1). Overexpression of CycC decreased the protein levels of SREBP-1a in the absence of insulin and blocked insulin-induced accumulation of SREBP-1a (Figure 7D). Furthermore, overexpression of CycC completely inhibited insulin-induced activation of the FAS promoter (Figure 7E). However, overexpression of wild-type CDK8 had little effect on the FAS promoter (data not shown). This is probably because increasing CDK8 alone is not sufficient for CDK8 kinase activity, substrate recognition, and/or localization in the absence of CycC. Finally, CycC overexpression in fat body of Drosophila larvae also inhibited re-feeding–induced upregulation of expression of lipogenic genes, such as dFAS (Figure 7F), and triglyceride accumulation (data not shown). Together, our data strongly support a functional role of CDK8-CycC in feeding/insulin-induced activation of nuclear SREBP-1c and de novo lipogenesis.
Discussion
In contrast to the sterol regulation of SREBP-2, insulin functions as the major activator of SREBP1C gene expression and inducer of SREBP-1c precursor processing to the mature nuclear form (5, 45–47). Numerous studies have established a key role of nuclear SREBP-1c on lipogenic gene expression and de novo lipogenesis (6, 16, 28, 44, 48). In this regard, SREBP-1c levels are elevated in mice and humans under a variety of pathophysiologic states, including obesity, insulin resistance associated with hyperinsulinemia, high carbohydrate diet, excessive alcohol consumption, and non-alcoholic fatty liver disease (NAFLD) — in the case of the latter being directly shown to increase the proportion of liver VLDL triglyceride production by de novo lipogenesis from 2%–5% to 20%–30% (41, 49–52). In those disease states, it is generally thought that the increased de novo lipogenesis results from high levels of insulin driving SREBP-1c transcription and precursor processing through PI3K- and mTORC1-dependent pathways, despite the marked insulin resistance to gluconeogenesis (14, 53, 54). However, these studies have only focused on the mechanisms of transcriptional activation of SREBP-1c expression and have not considered changes in the transcriptional activity or protein stability of nuclear SREBP-1c protein.
In this study, we identified a highly conserved role for CDK8 in the control of fatty acid and triglyceride metabolism. In Drosophila larvae, loss of CDK8 or CycC increased SREBP-dependent neutral lipid accumulation in fat body and lipogenic gene expression. Similarly, RNAi knockdown in hepatocytes in vitro or mouse liver in vivo also increased triglyceride levels, along with increasing SREBP target gene expression and de novo lipogenesis. Surprisingly, CycC and CDK8 levels were negatively regulated by food intake in mouse liver and by insulin in cultured cells. Thus, we propose a model whereby insulin stimulates de novo lipogenesis by downregulating the CDK8-CycC complex, which normally inhibits de novo lipogenesis through promoting nuclear SREBP-1c degradation. This new mechanism of insulin-induced lipogenesis is in addition to the well-documented function of insulin in stimulating SREBP-1c at the transcriptional level. Consistent with our model, overexpression of CycC blocks insulin-induced stabilization of nuclear SREBP-1 and lipogenic gene expression in vitro and in vivo. These observations establish an important and physiological role of CDK8-CycC in regulating fatty acid biosynthesis.
CDK8 and CycC are best known for their functions as the subunits of the Mediator complex, which serves as a transcription cofactor complex (26). Along with MED12 and MED13, they form the CDK8 submodule of the Mediator complex. However, unlike that of CDK8 and CycC, fat body–specific knockdown of MED12 and MED13 did not cause statistically significant changes in lipid levels in Drosophila larvae, although there was a trend toward a decrease in lipids in MED13-knockdown larvae (Supplemental Figure 5). Similarly, MED12 or MED13 knockdown had no effect on SREBP-1 in mammalian cells (data not shown). These results support a specific role of CDK8-CycC in lipid metabolism and are consistent with a previous report demonstrating functions for the CDK8-CycC complex distinct from those of the MED12-MED13 complex in regulating developmental patterns in Drosophila (23). Our data also support a model whereby specific subunits of the Mediator complex are involved in only specific biological pathways, although the Mediator complex collectively may regulate most of the RNAPII-controlled gene expression. Alternatively, CDK8-CycC may function independent of the Mediator complex. Mechanistically, CDK8-CycC inhibits lipid accumulation largely by repressing SREBP-mediated gene expression. Our biochemical analyses have revealed that SREBP-1c can be directly phosphorylated by CDK8 at a conserved T402 residue (for human SREBP-1c) in vitro and in cultured mammalian cells. Furthermore, we observed that phosphorylation of SREBP-1 at T402 promotes its degradation. Thus, this phosphorylation event provides a critical control mechanism to regulate the level of nuclear SREBP-1c protein by increasing ubiquitination and subsequent rate of degradation without affecting the levels of the precursor SREBP-1c protein (Figure 3A, Figure 4A, and Figure 6A). These results suggest a simple explanation for the increased fat accumulation on CDK8 or CycC mutants or in cells with reduced levels of CDK8 and CycC. That is, reduction of CDK8 or CycC will result in hypophosphorylation of SREBP-1, which increases the stability of the nuclear SREBP proteins, thereby allowing increased expression of SREBP target genes involved in de novo lipogenesis.
It has been known that phosphorylation of nuclear SREBP proteins controls their stability (35, 55). GSK-3β, which functions downstream of insulin signaling, was previously reported to be involved in phosphorylation of SREBP proteins at several conserved sites, including T426 of SREBP-1a (the site corresponding to T402 of SREBP-1c) (35, 55). In this study, we identified that CDK8 can directly phosphorylate the T402 site of SREBP-1c. It is not uncommon that the same threonine or serine residue of a protein can be phosphorylated by multiple kinases. The fact that the T402 site of SREBP-1c can be targeted by both GSK-3β and CDK8 is consistent with the key role of phosphorylation on this site for the binding of the E3 ligase SCFFbw7b (35, 55).
The importance of these findings is underscored by the following two points. First, the observed high rates of lipogenesis and enhanced SREBP-target gene expression in dyslipidemic states in CDK8 and CycC mutants cannot be solely accounted for by changes in SREBP-1c transcription or maturation. It will be important to determine whether the control over nuclear SREBP-1c protein levels through the CDK8-CycC complex can provide a mechanistic basis for the enigmatic enhancement of lipogenesis that occurs during insulin resistance and diabetes. Second, CDK8 has been recently identified as an oncoprotein in melanoma and colorectal cancers (56, 57). Aberrant lipid and carbohydrate metabolism is a universal feature of human cancer cells; however, the mechanisms linking such aberrant metabolism and tumorigenesis remain poorly understood. Our results suggest that dysregulation of CDK8 may not only promote tumorigenesis, but also disrupt the cellular lipid homeostasis by lifting its repressive effect on SREBP-dependent transcription. It will be important to investigate in the future whether such a mechanism is functional in different types of human cancer cells.
Methods
Drosophila stocks and genetics.
The w1118 and Oregon R Drosophila strains were used as controls. The null alleles of Cdk8 (w1118; +; FRT80B Cdk8K185/TM3 Sb) and CycC (w1118; +; FRT82B CycCY5/TM3 Sb) were provided by Henri-Marc Bourbon and Muriel Boube (Centre de Biologie du Développement, Université Paul Sabatier, Toulouse, France), and we replaced the balancer chromosome TM3 Sb with TM6B to facilitate the identification of homozygous mutant larvae. The fat body–specific Adh4-Gal4 and FB-Gal4 lines were obtained from Thomas Neufeld (Department of Genetics, University of Minnesota, Minneapolis, Minnesota, USA). The transgenic RNAi lines for Cdk8 and CycC were generated using the pVALIUM10 vector (58). Briefly, an 845-bp fragment of the dCDK8 gene and an 838-bp fragment of the dCycC gene were chosen using the online SnapDragon program developed by the Norbert Perrimon laboratory ( http://www.flyrnai.org/cgi-bin/RNAi_find_primers.pl). The PCR primers were: dCDK8 forward, 5′-CACCGAGCGTACGAAAGTGGAGGA-3′; dCDK8 reverse, 5′-AGCGAGCAGGTGGAGTATGT-3′; dCycC forward, 5′-CACCGGCAATTTTTGGCAGAGTTC-3′; and dCycC reverse, 5′-TGGGCAATACCAGCTAATGA-3′. The PCR fragments were cloned into entry vectors using a pENTR-D-TOPO cloning kit (Invitrogen), and the product was verified by diagnostic digestion. The fragments were then recombined into the pVALIUM10 vector, and the final constructs were verified by DNA sequencing. To knock down dSREBP (HLH106), we generated a miRNA transgenic line using the pVALIUM20 vector, which was shown to efficiently knock down target genes in both soma and germ line (59). The primers were: dSREBP-1F, 5′-CTAGCAGTCCGGTTATGCTTAGATTGTAATAGTTATATTCAAGCATATTACAATCTAAGCATAACCGGGCG-3′ and dSREBP-1R, 5′-AATTCGCCCGGTTATGCTTAGATTGTAATATGCTTGAATATAACTATTACAATCTAAGCATAACCGGACTG-3′. The final constructs were verified by DNA sequencing. UAS-CycC was constructed by inserting wild-type CycC cDNA (from Patrick O’Farrell, UCSF, San Francisco, California, USA) into the pTFHW vector (T. Murphy, Drosophila Genomic Resource Center, Bloomington, Indiana, USA), and the final construct was verified by sequencing. Transgenic flies were generated by injecting the vectors into early embryos (Genetic Services Inc). The transgenic RNAi lines were genetically combined with either Adh-Gal4 or FB-Gal4 using standard Drosophila genetics.
Microarray analyses.
Third instar larvae of Oregon R Drosophila (control), Cdk8-null (w1118; +; Cdk8K185), and CycC-null (w1118; +; CycCY5) were collected at the wandering stage. Total RNA was extracted from 10 larvae (triplicate for each genotype) with 1 ml of TRIzol Regent (Invitrogen) according to the manufacturer’s instructions. RNA quality was evaluated using the Agilent 2100 Bioanalyzer (Agilent). RNA (1 μg) was used for Affymetrix One-Cycle Target Labeling as described by the manufacturer (Affymetrix). Each of 6 Affymetrix Drosophila genome arrays was hybridized for 16–18 hours with biotin-labeled fragmented cRNA (10 μg) in 200 μl hybridization mixture according to the manufacturer’s protocol. Arrays were washed and stained using GeneChip Fluidic Station 450, and hybridization signals were amplified using antibody amplification with goat IgG (Sigma-Aldrich) and anti-streptavidin biotinylated antibody. Chips were scanned on a Affymetrix GeneChip Scanner 3000 using Affymetrix Command Console Software. Raw data were normalized using Robust Multichip Average and normalized to control samples with GeneSpring GX 11.0.2 software (Agilent). A volcano plot was used to identify differentially expressed genes using parametric testing assuming equal variances and no multiple testing corrections. Pathway analysis of differentially expressed genes was performed employing Ingenuity software (Ingenuity Systems). The complete sets of microarray data have been deposited in the ArrayExpress database ( http://www.ebi.ac.uk/arrayexpress/; accession number E-MTAB-1066).
Oil red O staining and quantification.
Drosophila larvae were partially dissected in 1× PBS and then fixed in 4% formalin in 1× PBS for 15 minutes at room temperature. After three quick rinses with distilled water, the female larvae were stained with 5 ml of 0.036% oil red O (by mixing 6 ml of 0.1% oil red O in isopropanol with 10.5 ml distilled water, then filtering through a 0.45-nm filter; Millipore) for 25 minutes at room temperature. After 2 quick rinses (1 minute for each) with 70% isopropanol and 1 quick rinse with distilled water, each larva was transferred into individual Eppendorf tubes and dried overnight. The oil red O from each larva was then extracted by adding 0.3 ml isopropanol and rocking for 8 hours before measuring the O.D. at 510 nm. The amount of oil red O staining was normalized to the control, and the results of at least 3 independent experiments were combined and presented. Slides of mouse livers were stained with oil red O similarly.
Antibodies.
Anti-CDK8 (Abcam and Santa Cruz Biotechnology Inc.), anti–cyclin C (Invitrogen), anti–SREBP-1 (Santa Cruz Biotechnology Inc.), anti-FAS (Cell Signaling Technology), anti-Flag (Sigma-Aldrich), anti-HA (Covance), anti–p-TP (Cell Signaling Technology), anti–p-S (Invitrogen), anti–β-tubulin (Invitrogen), and anti-TBP (Fisher Scientific) antibodies were used in this study.
Tissue culture.
HEK293, HepG2, and FAO cells were purchased from ATCC and cultured in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone), 2 mM l-glutamine (Gibco), 100 U/ml penicillin (Gibco), and 100 μg/ml streptomycin (Gibco) at 37°C under humidified air containing 5% CO2. Primary rat hepatocytes were isolated by perfusion of rat livers by David Neufeld (Marion Bessin Liver Research Center, Albert Einstein College of Medicine). For all perfusions, Chee medium (pH 7.2) was supplemented with 10 mM HEPES. Washout medium was further supplemented with heparin (2.0 U/ml) and EGTA (0.5 mM), and digestion medium was supplemented with 500 mg/l collagenase. Viable primary rat hepatocytes were enriched by low-speed centrifugation (500 g) for 3 minutes. Typically, the yield of isolated hepatocytes was approximately 3 × 107 cells with a viability of greater than 80% as determined by trypan blue dye exclusion. The primary rat hepatocytes were further selected for experiments by seeding 1.5 × 106 cells on a 6-cm BD BioCoat dish (BD Biosciences) supplemented with low-glucose DMEM (Gibco), 5% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. After approximately 4 hours of incubation, non-adherent cells were washed twice with 1× PBS, and the resulting hepatocytes were cultured in the above DMEM medium for further treatment. For insulin treatment, cells were fasted in serum-free medium for 4 hours.
Preparation of lentivirus.
pGIPZ-shRNA plasmids were purchased from Thermo Scientific. Lentivirus was packaged by the TransLenti Viral GIPZ Packaging System (Thermo Scientific). The day before transfection of pGIPZ plasmids, HEK293T cells were plated at a density of 5.5 × 106 cells per 100-mm plate. For each plate, 9 μg plasmids and 28.5 μg viral packaging mix were transfected into HEK293T cells by Arrest-In transfection reagent. Fiver hours after transfection, culture medium was replaced with regular DMEM. Transfection efficiency was then examined two days after transfection by the percentage of GFP-positive cells. Culture medium was collected at day 3 and centrifuged at 2,000 g for 20 minutes at 4°C. The virus-containing supernatant was kept at –80°C in aliquots.
Protein extraction and immunoblotting.
For whole cell extracts, cells or homogenized mouse tissues were lysed in a buffer containing 50 mM Tris-HCl (pH 8.0), 0.1 mM EDTA, 420 mM NaCl, 0.5% NP-40, 0.05% SDS, 10% glycerol, 1 mM dithiothreitol, 2.5 mM PMSF, 1 mM benzamidine, 1 mg/l aprotinin, and 0.1 mM of the calpain inhibitor ALLN (Santa Cruz Biotechnology Inc.). Supernatants were collected after centrifugation at 20,000 g for 20 minutes at 4°C. For nuclear extracts, cells or mouse tissues were first suspended in 5 volumes of buffer A (10 mM Tris-HCl pH 8.0, 1.5 mM MgCl2, 10 mM KCl, 1 mM dithiothreitol, 2.5 mM PMSF, 1 mM benzamidine, 1 mg/l aprotinin), homogenized in a glass homogenizer until approximately 90% cells were broken, and centrifuged at 2,000 g for 5 minutes at 4°C. The pellet was washed once with buffer A and resuspended in buffer C (20 mM Tris-HCl pH 8.0, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 20% glycerol, 12% sucrose, 1 mM dithiothreitol, 2.5 mM PMSF, 1 mM benzamidine, and 1 mg/l aprotinin). This suspension of nuclei was rotated at 4°C for 40 minutes and centrifuged 20,000 g for 20 minutes at 4°C. Supernatant was dialyzed against 500 volumes of buffer D (20 mM Tris-HCl pH 8.0, 100 mM KCl, 1.5 mM MgCl2, 0.1 mM EDTA, 20% glycerol, 1 mM dithiothreitol, 2.5 mM PMSF, and 1 mM benzamidine). After centrifugation at 20,000 g for 20 minutes at 4°C, the resulting supernatant was designated as nuclear extract. Protein concentrations were measured with a BCA kit (Pierce). A given amount of whole cell extract or nuclear extract was mixed with 5× SDS loading buffer (0.25 M Tris-HCl pH 6.8, 10% SDS, 50% glycerol, 0.05% bromphenol blue, 500 mM dithiothreitol). After boiling for 3 minutes, the proteins were resolved by NuPAGE 4%–12% Bis-Tris gel (Invitrogen) and transferred to nitrocellulose or PVDF membrane by iBlot Gel Transfer Kit (Invitrogen). After blocking in 5% nonfat milk in 1× TBST, the membrane was incubated with specific primary antibodies with appropriate dilution for 2 hours at room temperature and washed 3 times with 1× TBST (10 minutes each). Then, the membrane was incubated with the HRP-conjugated secondary antibodies (1:10,000 dilution) for 1 hour at room temperature. After washing 3 times with 1× TBST (10 minutes each), the HRP signals were visualized by the SuperSignal West Pico kit (Pierce) according to the manufacturer’s instructions.
In vitro kinase assay.
Endogenous CDK8 proteins were immunoprecipitated from HEK293 cell nuclear extracts with anti-CDK8 antibody on protein A/G sepharose beads. Flag-tagged CDK8 proteins expressed in HEK293 cells by transient transfection were immunoprecipitated using anti-Flag M2 affinity gel (Sigma-Aldrich). After incubation for 2 hours at 4°C, protein A/G sepharose or anti-Flag beads were washed 3 times with IP buffer and mixed with GST proteins. The mixture was washed once with the kinase buffer (50 mM Tris-HCl pH 7.4, 50 mM NaCl, 10 mM MgCl2, 1 mM MnCl2 and 10% glycerol) and incubated in 50 μl of kinase buffer containing 50 mM ATP for 1 hour at 30°C. The reaction was terminated by boiling in 5× SDS loading buffer. The phosphorylation signals were detected using specific anti–p-TP and anti–p-S antibodies.
RNA preparation and quantitative PCR analysis.
Total RNA was isolated from cells and mouse livers using the TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. RNA concentration was measured by a NanoDrop spectrophotometer (Thermo). After removing genomic DNA with RQ1 RNase-free DNase I (Promega), cDNA was synthesized by a First-Strand cDNA Synthesis Kit (GE Healthcare). Synthesized cDNA was diluted for real-time PCR. Each real-time PCR reaction mixture contained 10 μl FastStart universal SYBR Green Master mix (Roche), 1 μl primer (250 nM each), and 9 μl diluted cDNA. Real-time PCR was performed using the StepOnePlus Real-Time PCR System (Applied Biosystems). The cycling parameters consisted of 95°C incubation for 10 minutes for enzyme activation and DNA denaturation, followed by 40 PCR amplification cycles consisting of 95°C for 15 seconds and 60°C for 1 minute. The thermocycling program was followed by a melting program of 95°C for 15 seconds (denaturation), 60°C for 1 minute (annealing), and then 60–95°C at a transition rate of 0.3°C/s with continual monitoring of fluorescence. Data analysis was performed by software provided with the StepOnePlus Real-Time PCR System. PCR primers are listed in Supplemental Table 2.
Mouse treatment.
Male C57BL/6J mice purchased from The Jackson Laboratory were maintained under a 12-hour dark cycle with free access to water and standard mouse diet (10% calories from fat). For transient deletion of hepatic CDK8, 8- to 10-week-old mice were injected with adenovirus (~1.5 × 109 infectious particles per mouse) expressing nonspecific shRNA (control) or CDK8 shRNA (Welgen Inc.) through tail veins. For de novo lipogenesis experiments, 9 days after viral injection, food was removed in the morning and mice received an i.p. injection of deuterated water (2H2O) containing 0.9% sodium chloride at a concentration of approximately 4% lean body mass. Mice were provided drinking water containing 4% 2H2O. Five hours later, mice were sacrificed, and pieces of liver tissues and plasma were sent to the Metabolic Center at Albert Einstein College of Medicine to determine the palmitate levels by gas chromatography and mass spectrometry.
Metabolites.
Plasma triglyceride, glycerol, and non-esterfied fatty acid (NEFA) levels were measured using Infinity Triglycerides Reagent (Thermo Scientific), free glycerol reagent (Sigma-Aldrich), and HR Series NEFA-HR (Wako). Liver triglyceride levels were analyzed by an Adipogenesis Assay Kit (Biovision) and normalized by total protein levels.
Statistics.
For quantitative measurement of mRNA, protein, triglycerides, oil red O staining, NEFA, glycerol, and insulin, the comparison of 2 different groups was carried out using 2-tailed unpaired Student’s t test. Results are presented as mean ± SD. Differences was considered statistically significant when P was less than 0.05.
Study approval.
All experiments using mice and rats conformed to protocols approved by the Animal Care and Use Committee of Albert Einstein College of Medicine.
Supplementary Material
Acknowledgments
We thank Henri-Marc Bourbon and Muriel Boube for dCDK8 and dCycC mutant flies and the Bloomington Drosophila Stock Center for other fly strains. We are grateful to Liz Perkins, Norbert Perrimon, Laura Holderbaum, and the TRiP at Harvard Medical School (GM084947), as well as Patrick O’Farrell and Terry Orr-Weaver for providing transgenic RNAi fly stocks and reagents used in this study. We thank Ana Maria Cuervo, Craig Kaplan, Geoffrey Kapler, E. Richard Stanley, and Liang Zhu for critical reading of the manuscript. We regret the inability to cite many original studies owing to space limitations. This work was supported by grants from the Diabetes Research and Training Program (P60-DK020541), the American Diabetes Association (7-11-BS-173), and the NIH (DK093623) to F. Yang; and a grant from the American Heart Association to J.-Y. Ji.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(7):2417–2427. doi:10.1172/JCI61462.
Jie Zhou’s present address is: Department of Endocrinology and Metabolism, Xijing Hospital, the Fourth Military Medical University, Xi’an, China.
References
- 1.van Herpen NA, Schrauwen-Hinderling VB. Lipid accumulation in non-adipose tissue and lipotoxicity. Physiol Behav. 2008;94(2):231–241. doi: 10.1016/j.physbeh.2007.11.049. [DOI] [PubMed] [Google Scholar]
- 2.Muoio DM, Newgard CB. Obesity-related derangements in metabolic regulation. Annu Rev Biochem. 2006;75:367–401. doi: 10.1146/annurev.biochem.75.103004.142512. [DOI] [PubMed] [Google Scholar]
- 3.Cheung O, Sanyal AJ. Abnormalities of lipid metabolism in nonalcoholic fatty liver disease. Semin Liver Dis. 2008;28(4):351–359. doi: 10.1055/s-0028-1091979. [DOI] [PubMed] [Google Scholar]
- 4.Reaven GM. Insulin resistance: the link between obesity and cardiovascular disease. Med Clin North Am. 2011;95(5):875–892. doi: 10.1016/j.mcna.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Shimomura I, Bashmakov Y, Ikemoto S, Horton JD, Brown MS, Goldstein JL. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci U S A. 1999;96(24):13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimano H, et al. Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes. J Biol Chem. 1999;274(50):35832–35839. doi: 10.1074/jbc.274.50.35832. [DOI] [PubMed] [Google Scholar]
- 7.Liang G, Yang J, Horton JD, Hammer RE, Goldstein JL, Brown MS. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J Biol Chem. 2002;277(11):9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- 8.Amemiya-Kudo M, et al. Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes. J Lipid Res. 2002;43(8):1220–1235. [PubMed] [Google Scholar]
- 9.Jeon T-I, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol Metab. 2012;23(2):65–72. doi: 10.1016/j.tem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it’s been. Genes Dev. 2009;23(22):2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hua X, et al. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc Natl Acad Sci U S A. 1993;90(24):11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yokoyama C, et al. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75(1):187–197. [PubMed] [Google Scholar]
- 13.Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77(1):53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 14.Ferre P, Foufelle F. SREBP-1c transcription factor and lipid homeostasis: clinical perspective. Horm Res. 2007;68(2):72–82. doi: 10.1159/000100426. [DOI] [PubMed] [Google Scholar]
- 15.Hua X, Wu J, Goldstein JL, Brown MS, Hobbs HH. Structure of the human gene encoding sterol regulatory element binding protein-1 (SREBF1) and localization of SREBF1 and SREBF2 to chromosomes 17p11.2 and 22q13. Genomics. 1995;25(3):667–673. doi: 10.1016/0888-7543(95)80009-B. [DOI] [PubMed] [Google Scholar]
- 16.Shimano H, Horton JD, Shimomura I, Hammer RE, Brown MS, Goldstein JL. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest. 1997;99(5):846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest. 1997;99(5):838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naar AM, Beaurang PA, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398(6730):828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 19.Taatjes DJ, Naar AM, Andel F 3rd, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295(5557):1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- 20.Xu W, Ji JY. Dysregulation of CDK8 and Cyclin C in tumorigenesis. J Genet Genomics. 2011;38(10):439–452. doi: 10.1016/j.jgg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conaway RC, Conaway JW. Origins and activity of the Mediator complex. Semin Cell Dev Biol. 2011;22(7):729–734. doi: 10.1016/j.semcdb.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2011;11(11):761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loncle N, et al. Distinct roles for Mediator Cdk8 module subunits in Drosophila development. EMBO J. 2007;26(4):1045–1054. doi: 10.1038/sj.emboj.7601566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol Cell Biol. 2009;29(3):650–661. doi: 10.1128/MCB.00993-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci. 2005;30(5):250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35(6):315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris EJ, et al. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455(7212):552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horton JD, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A. 2003;100(21):12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rome S, et al. Microarray analyses of SREBP-1a and SREBP-1c target genes identify new regulatory pathways in muscle. Physiol Genomics. 2008;34(3):327–337. doi: 10.1152/physiolgenomics.90211.2008. [DOI] [PubMed] [Google Scholar]
- 30.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100(4):391–398. doi: 10.1016/S0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 31.Rawson RB. The SREBP pathway — insights from Insigs and insects. Nat Rev Mol Cell Biol. 2003;4(8):631–640. doi: 10.1038/nrm1174. [DOI] [PubMed] [Google Scholar]
- 32.Yang F, et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442(7103):700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 33.Leclerc V, Tassan JP, O’Farrell PH, Nigg EA, Leopold P. Drosophila Cdk8, a kinase partner of cyclin C that interacts with the large subunit of RNA polymerase II. Mol Biol Cell. 1996;7(4):505–513. doi: 10.1091/mbc.7.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang F, DeBeaumont R, Zhou S, Naar AM. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc Natl Acad Sci U S A. 2004;101(8):2339–2344. doi: 10.1073/pnas.0308676100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sundqvist A, et al. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7). Cell Metab. 2005;1(6):379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 36.Rickert P, Seghezzi W, Shanahan F, Cho H, Lees E. Cyclin C/CDK8 is a novel CTD kinase associated with RNA polymerase II. Oncogene. 1996;12(12):2631–2640. [PubMed] [Google Scholar]
- 37.Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell. 2004;16(4):509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 38.Alarcon C, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139(4):757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramanathan Y, et al. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J Biol Chem. 2001;276(14):10913–10920. doi: 10.1074/jbc.M010975200. [DOI] [PubMed] [Google Scholar]
- 40.Westerling T, Kuuluvainen E, Makela TP. Cdk8 is essential for preimplantation mouse development. Mol Cell Biol. 2007;27(17):6177–6182. doi: 10.1128/MCB.01302-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoeckman AK, Towle HC. The role of SREBP-1c in nutritional regulation of lipogenic enzyme gene expression. J Biol Chem. 2002;277(30):27029–27035. doi: 10.1074/jbc.M202638200. [DOI] [PubMed] [Google Scholar]
- 42.Ishii S, Iizuka K, Miller BC, Uyeda K. Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc Natl Acad Sci U S A. 2004;101(44):15597–15602. doi: 10.1073/pnas.0405238101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci U S A. 2004;101(19):7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci U S A. 1998;95(11):5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dif N, Euthine V, Gonnet E, Laville M, Vidal H, Lefai E. Insulin activates human sterol-regulatory-element-binding protein-1c (SREBP-1c) promoter through SRE motifs. Biochem J. 2006;400(1):179–188. doi: 10.1042/BJ20060499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yabe D, Komuro R, Liang G, Goldstein JL, Brown MS. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc Natl Acad Sci U S A. 2003;100(6):3155–3160. doi: 10.1073/pnas.0130116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yellaturu CR, Deng X, Park EA, Raghow R, Elam MB. Insulin enhances the biogenesis of nuclear sterol regulatory element-binding protein (SREBP)-1c by posttranscriptional down-regulation of Insig-2A and its dissociation from SREBP cleavage-activating protein (SCAP).SREBP-1c complex. J Biol Chem. 2009;284(46):31726–31734. doi: 10.1074/jbc.M109.050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsumoto E, et al. Time of day and nutrients in feeding govern daily expression rhythms of the gene for sterol regulatory element-binding protein (SREBP)-1 in the mouse liver. J Biol Chem. 2010;285(43):33028–33036. doi: 10.1074/jbc.M109.089391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimomura I, Bashmakov Y, Horton JD. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J Biol Chem. 1999;274(42):30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- 50.Aragno M, et al. SREBP-1c in nonalcoholic fatty liver disease induced by Western-type high-fat diet plus fructose in rats. Free Radic Biol Med. 2009;47(7):1067–1074. doi: 10.1016/j.freeradbiomed.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 51.You M, Fischer M, Deeg MA, Crabb DW. Ethanol induces fatty acid synthesis pathways by activation of sterol regulatory element-binding protein (SREBP). J Biol Chem. 2002;277(32):29342–29347. doi: 10.1074/jbc.M202411200. [DOI] [PubMed] [Google Scholar]
- 52.Donnelly KL, Smith CI, Schwarzenberg SJ, Jessurun J, Boldt MD, Parks EJ. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J Clin Invest. 2005;115(5):1343–1351. doi: 10.1172/JCI23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Curr Biol. 2009;19(22):R1046–R1052. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Postic C, Girard J. The role of the lipogenic pathway in the development of hepatic steatosis. Diabetes Metab. 2008;34(6 pt 2):643–648. doi: 10.1016/S1262-3636(08)74599-3. [DOI] [PubMed] [Google Scholar]
- 55.Bengoechea-Alonso MT, Ericsson J. A phosphorylation cascade controls the degradation of active SREBP1. J Biol Chem. 2009;284(9):5885–5895. doi: 10.1074/jbc.M807906200. [DOI] [PubMed] [Google Scholar]
- 56.Firestein R, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455(7212):547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kapoor A, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468(7327):1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ni JQ, et al. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods. 2008;5(1):49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ni JQ, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8(5):405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.