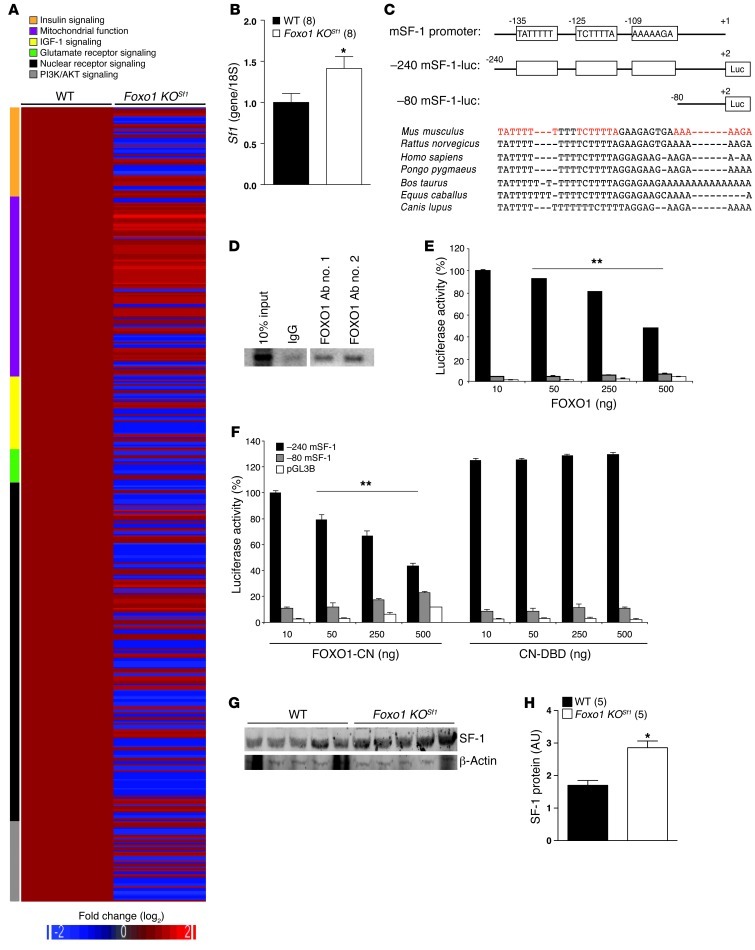
Figure 6. Transcriptional regulation of FOXO1 in the VMH.
(A) Heat map demonstrating gene expression profiling in the VMH of WT and Foxo1 KOSf1 mice. Relative fold changes of genes involved in indicated signaling pathways are shown (WT value = 1). (B) Sf1 mRNA levels in the VMH of 24-hour–fasted WT and Foxo1 KOSf1 mice (*P < 0.05, t test). (C) Schematic diagram of the 5′-flanking region of the mouse Sf1 gene, including the location of potential FOXO1 binding sites (white boxes). Shown below are the 5′-deletion constructs. Genomic alignments of SF-1 5′-flanking region of mammals (around FOXO1 putative binding sites). Red indicates the putative FOXO1 binding sites. (D) ChIP analysis showing direct binding of FOXO1 on the SF-1 promoter region in the VMH. (E) SF-1 promoter activity is significantly repressed by FOXO1 in Neuro2A cells in a dose-dependent manner (**P < 0.0001, only for –240 mSF-1, 1-way ANOVA). (F) SF-1 promoter activity is significantly repressed by a constitutive nuclear active form of FOXO1 (FOXO1-CN) in a dose-dependent manner but not by a DNA-binding deletion mutant of FOXO1 (CN-DBD) in Neuro2A cells (**P < 0.0001, only for –240 mSF-1, 1-way ANOVA). (G and H) SF-1 protein levels in 24-hour–fasted WT and Foxo1 KOSf1 animals (*P < 0.05, t test). The data are expressed as mean ± SEM (B, E, F, and H).