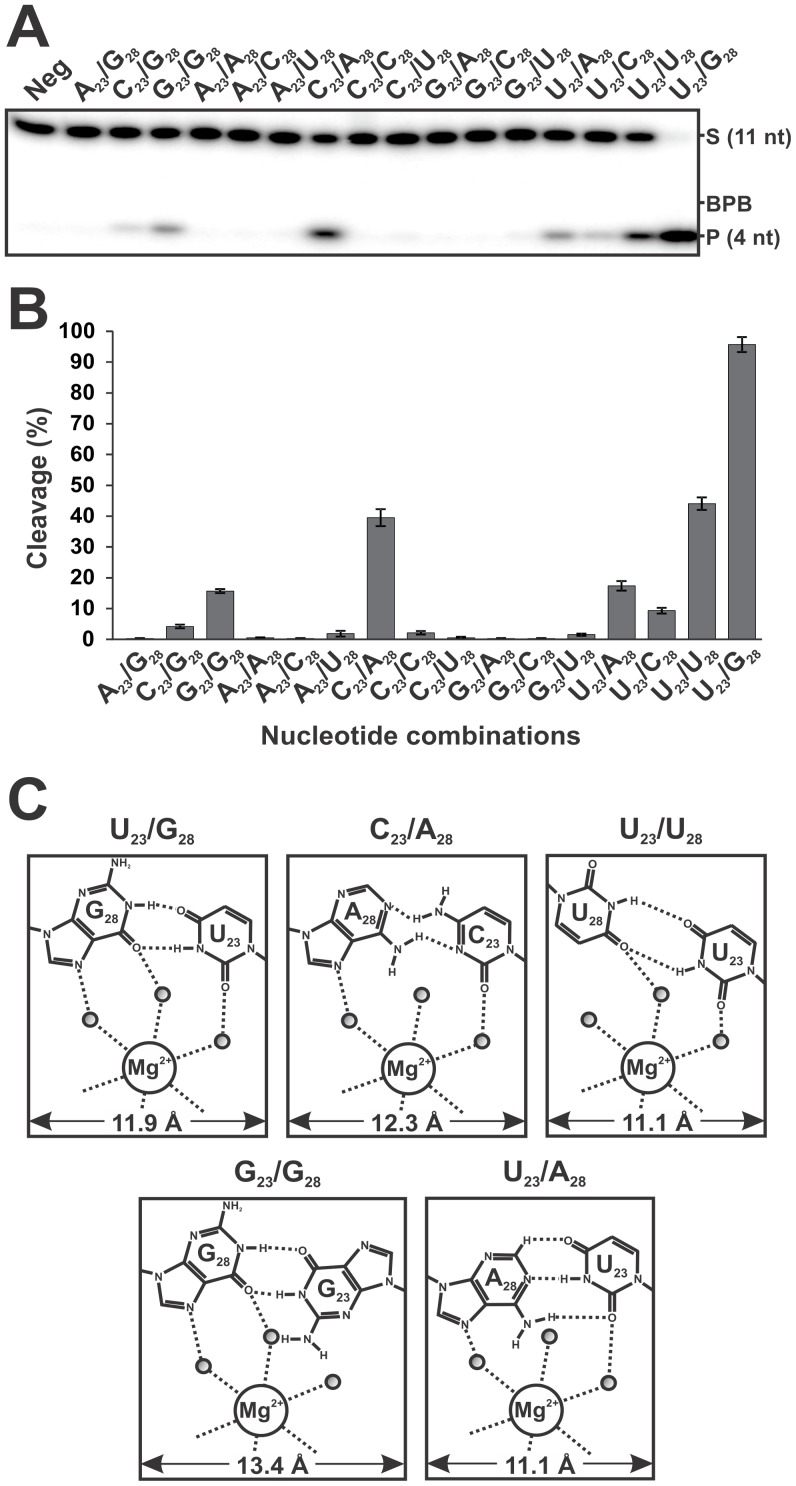
Figure 2. Cleavage activities of all of the ribozymes mutated in positions 23 and 28.
(A) Autoradiogram of the denaturing PAGE gel used for the analysis of the cleavage reactions. In all cases, the ribozymes (100 nM) were incubated for 2 h with trace amounts of 5′-end-labeled substrate (<1 nM). The positions of the bromophenol blue dye (BPB), the 11-nt substrate (S) and the 4-nt product (P) are indicated. “Neg” represents a cleavage reaction performed in the absence of any Rz. (B) Graphical representation of the cleavage percentages for all of the 16 nucleotide combinations examined. The values are means of at least 2 different experiments and the error bars represent the standard deviation (C) Putative H-bond representation and magnesium ion binding site of the wild-type (U23/G28) and four other nucleotide combinations.