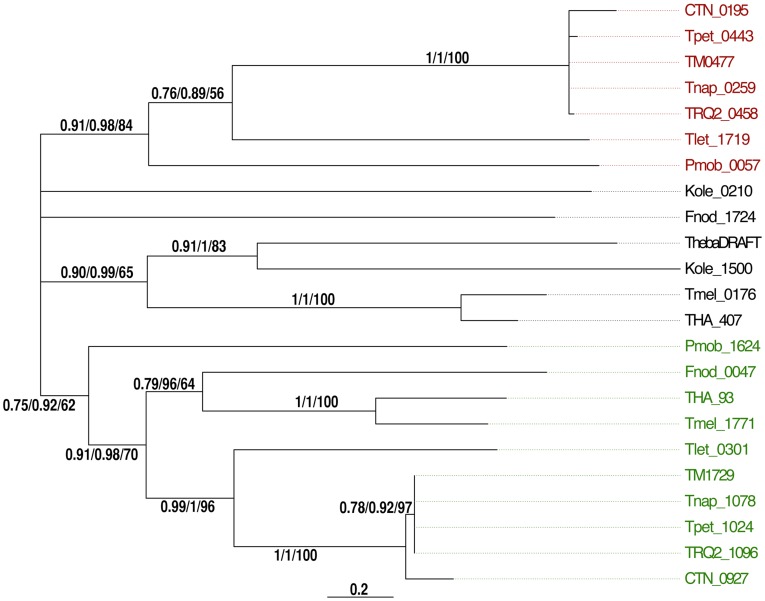
Figure 2. Maximum likelihood phylogenetic tree of OmpA protein sequences from several Thermotogales species.
Red labels indicate likely T. maritima OmpA1 (TM0477) orthologs, green labels indicate likely OmpA2 (TM1729) orthologs, and homologs whose type of homology cannot be ascertained are labeled in black. The tree was calculated as unrooted phylogeny, but is depicted as rooted between the likely OmpA1 and OmpA2 clusters. Branches with approximate Likelihood Ratio Test (aLRT) support values ≤0.75 were collapsed. aLRT, posterior probability, and bootstrap support values are given above or below the branch to which they pertain. Organism abbreviations and gene identification numbers for loci are Petrotoga mobilis (Pmob_0057, 160901548 and Pmob_1624, 160903060); T. lettingae (Tlet_1719, 157364570 and Tlet_0301, 157363168); T. petrophila (Tpet_1024, 148270158 and Tpet_0443, 148269583); T. maritima (TM0477, 15643243 and TM1729, 15644475); T. napthophila (Tnap_1078, 281412500 and Tnap_0259, 281411698) T. neapolitana (CTN_0195, 222099169 and CTN_0927, 222099901); Thermotoga. sp. strain RQ2 (TRQ2_0458, 170288259 and TRQ2_1096, 170288887); Fervidobacterium nodosum (Fnod_1724, 154250391 and Fnod_0047, 154248750); Kosmotoga olearia (Kole_0210, 239616617 and Kole_1500, 239617873); Thermosipho africanus (THA_407, 217076525 and THA_93, 217076226); Thermosipho melanesiensis (Tmel_0176, 150020084 and Tmel_1771, 150021641); Thermotogales bacterium mesG.Ag.4 (Mesotoga prima) (ThebaDRAFT_0522, 307297745; now Theba_0318).