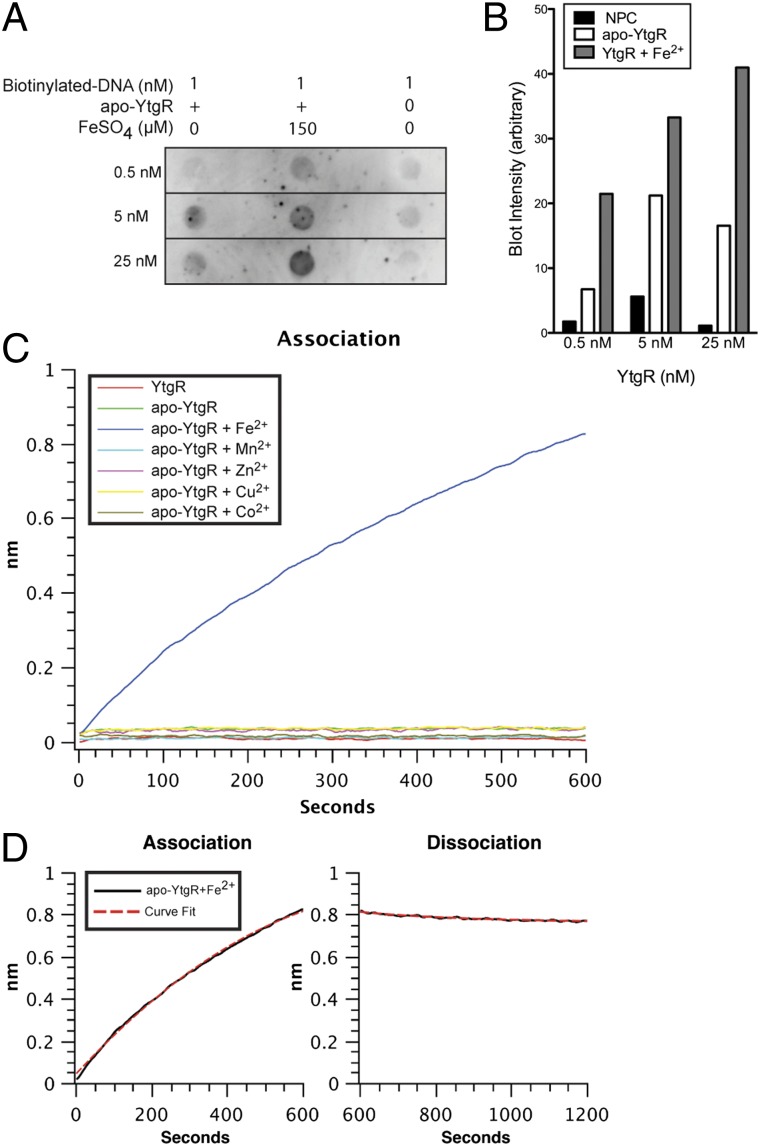
Fig. 2.
Recombinant version of the YtgR domain is a functional, iron-dependent DNA-binding polypeptide in vitro. (A) A labeled DNA molecule corresponding to the region upstream of ytgA was added to a solution containing various concentrations of apo-YtgR, in the presence or absence of iron. Blot represents one representative experiment of three. (B) Blot intensities were measured using ImageJ software, and the background level of an unused dot was subtracted from the measured values. Detection intensity increased within each apo-YtgR concentration upon supplementation of iron. (C) Real-time accumulation of biolayer was measured at the surface of an optical sensor that had been preloaded with a DNA oligonucleotide (corresponding to the region upstream of the ytgA coding sequence) and incubated in a solution containing purified YtgR or apo-YtgR in the presence of various divalent metals. Only the apo-YtgR plus Fe2+ reaction exhibited appreciable protein to DNA interaction. After allowing samples to associate with the sensor for 10 min, sensors were incubated in YtgR-free buffer and the dissociation was measured. (D) Rates of association (Kon) and dissociation (Koff) were determined by the “association-then-dissociation” function of the GraphPad Prism software platform for the apo-YtgR + Fe2+ binding reaction. A mean average KD of 3.45 × 10−8 M was measured over three independent experiments.