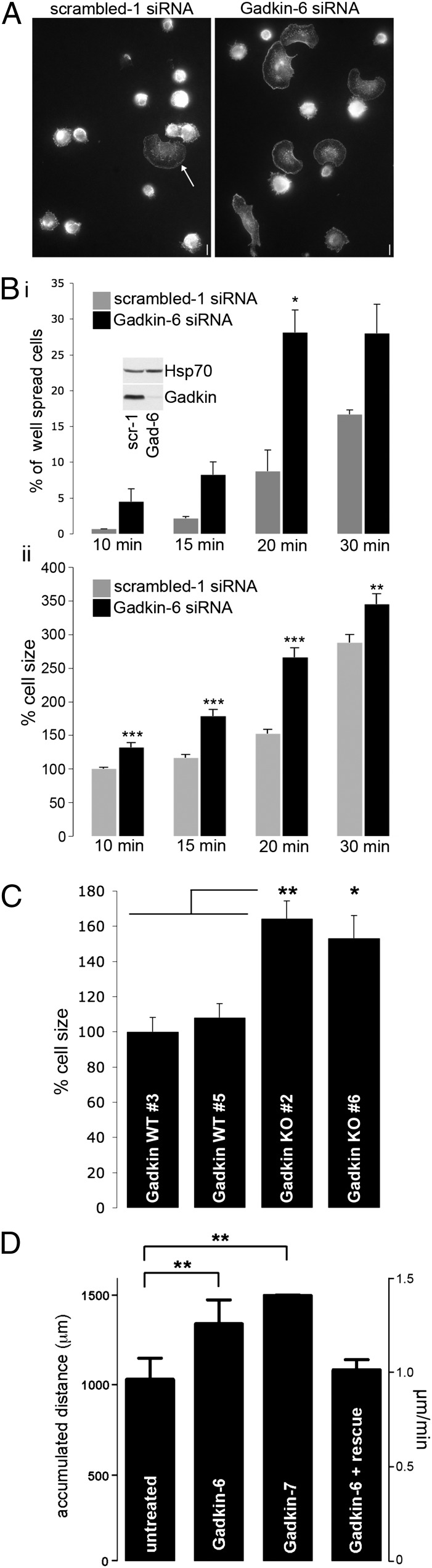
Fig. 1.
Gadkin negatively regulates cell spreading and migration. (A and B) B16F1 cells treated with scrambled or Gadkin-targeting siRNAs were trypsinized and seeded onto fibronectin-coated glass coverslips. After the indicated time intervals, cells were rapidly washed, fixed, and stained with phalloidin. (A) Representative images of control and Gadkin-depleted cells 20 min postplating. (Scale bar: 10 μm.) (Bi) Percentage of spread cells at different time points (two experiments; n = 128–299 cells per condition per experiment; *P < 0.05; unpaired Student’s t test). Symmetrically flattened out cells with a smooth plasmalemmal rim (see arrow in A) were classified as well spread. (Inset) Knockdown verified by immunoblot analysis. (Bii) Percentage cell size evaluated for the same experiment normalized to the size of control cells at 10 min (***P < 0.0001; unpaired Student’s t test). (C) Percentage cell size of two independent Gadkin WT and KO MEF cell lines 20–30 min postplating normalized to the size of Gadkin WT no. 3 cells (two experiments; n = 221–293 cells per cell line; *P < 0.01, **P < 0.001; one-way ANOVA plus Tukey’s post test). (D) B16F1 cells treated with Gadkin-targeting siRNAs and control cells were seeded onto laminin. Analysis of time lapse movies revealed that Gadkin-depleted cells migrate more, an effect that can be rescued by overexpression of Gadkin-WT-EGFP (n = 2–3; n = 30 cells per experiment; **P < 0.001; unpaired Student’s t test). Error bars always represent SEM.