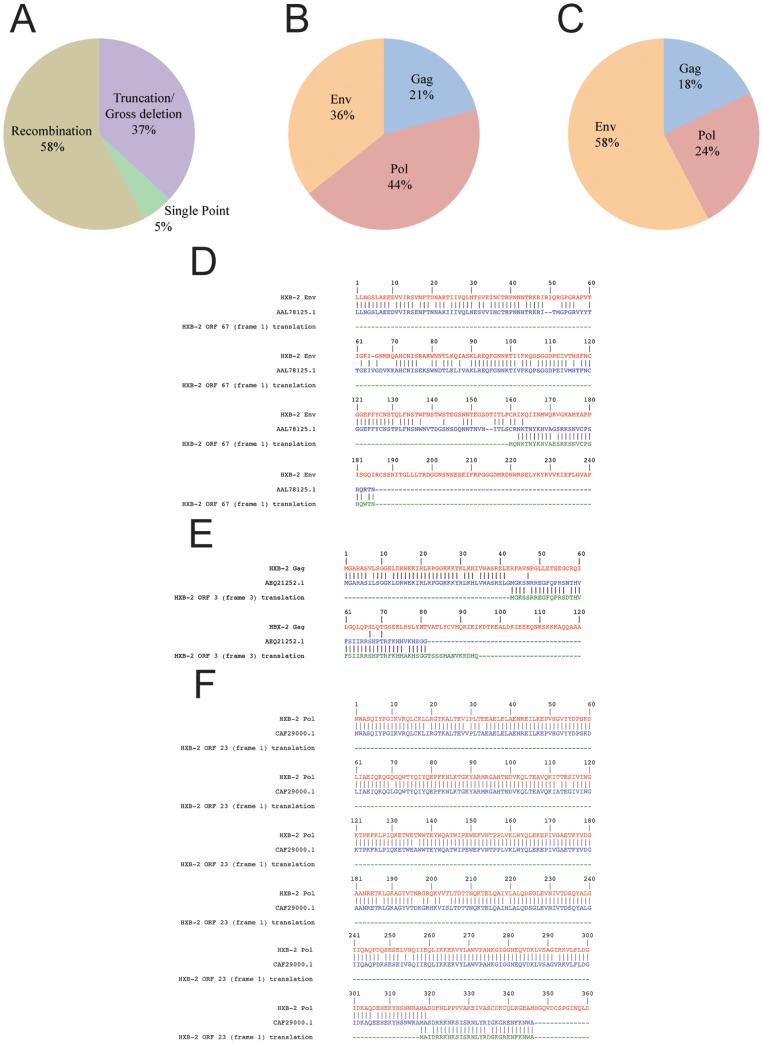
Figure 2. Alternate reading frame-encoded amino acids in circulating viral sequences.
A) Distribution of known mutant origins across viral ARF sequences attributable to a particular cause based on information associated with the sequence accession the NCBI nr protein database presented in Table S2. B) Composition of viral coding sequence computed as a percentage assigned to the coding sequence for the Env, Gag, Pol poly-proteins based on nucleotide base counts for a particular gene region compared to the total nucleotide count for the structural genes of the virus. Gag comprises 1,503 nt of the total of 6,878 nt of structural gene sequence; Pol comprises 3,139 nt of the total; Env comprises of 2,571 nt of the total. This is the distribution of origins within the genome that would be expected if originating events for the incorporation of ARF and their detection in circulating HIV-1 viral sequences were distributed randomly throughout the genome. C) The distribution of ARF incorporated into circulating viral sequences that was observed in our searches of NCBI nr protein database for ARF sequences in circulating HIV-1 viral sequences. The percentages were computed by dividing the number of BLAST hits with ARF sequence incorporated into a given gene region by the 123 total hits examined. D) A three-way alignment between the HXB-2 reference sequence for the Env region, the accession AAL78125.1 and the alternate reading frame encoded ORF 67. E) A three-way alignment between the HXB-2 reference sequence for the Gag region, the accession AEQ21252.1 and the alternate reading frame encoded ORF 3. F) A three-way alignment between the HXB-2 reference sequence for the Pol region, the accession CAF29000.1 and the alternate reading frame encoded ORF 23. All three-way alignments were generated by combining two pair-wise alignments created in Geneious, followed by manual editing. Note each accession is similar to both the HXB-2 reference sequence for the structural proteins and the alternate reading frame encoded sequence, but not to both sequences simultaneously within the same region of the sequence.